Advancing biological fluorescence microscopy with deep learning: a bibliometric perspective
Li Fu ,Cheng-Liang Yin ,Jia-Lei Wang ,Xiao-Zhu Liu
1College of Materials and Environmental Engineering,Hangzhou Dianzi University,Hangzhou 310018,China.2Medical Big Data Research Center,Medical Innovation Research Division of PLA General Hospital,Beijing 100853,China.3National Engineering Laboratory for Medical Big Data Application Technology,Chinese PLA General Hospital,Beijing 100853,China.4Department of Algorithm,Shining3D Technology Company Limitid,Hangzhou 311258,China.5Department of Cardiology,The Second Affiliated Hospital of Chongqing Medical University,Chongqing 400010,China.
Abstract Background: Fluorescence microscopy has increasingly promising applications in life science.This bibliometrics-based review focuses on deep learning assisted fluorescence microscopy imaging techniques.Methods:Papers on this topic retrieved by Core Collection on Web of Science between 2017 and July 2022 were used for the analysis.In addition to presenting the representative papers that have received the most attention,the process of development of the topic,the structure of authors and institutions,the selection of journals,and the keywords are analyzed in detail in this review.Results:The analysis found that this topic gained immediate popularity among scholars from its emergence in 2017,gaining explosive growth within three years.This phenomenon is because deep learning techniques that have been well established in other fields can be migrated to the analysis of fluorescence micrographs.From 2020 onwards,this topic tapers off but has attracted a few stable research groups to tackle the remaining challenges.Although this topic has been very popular,it has not attracted scientists from all over the world.The USA,China,Germany,and the UK are the key players in this topic.Keyword analysis and clustering are applied to understand the different focuses on this topic.Conclusion:Based on the bibliometric analysis,the current state of this topic to date and future perspectives are summarized at the end.
Keywords: deep learning;fluorescence microscopy;signal-to-noise ratio;convolutional neural network;cell segmentation
Introduction
In recent years,optical microscopic imaging techniques have developed rapidly,among which fluorescence microscopy has become an indispensable tool for studying the dynamic properties of cells,tissues,and organisms in time and space.Compared to electron microscopy,optical microscopy offers the advantage of low-or no-damage imaging,providing multidimensional structural and functional information about the sample without invading it [1].Today,fluorescence microscopes have developed many types,such as wide-field microscopes,confocal microscopes,two-photon microscopes,light-sheet microscopes,and super-resolution microscopes.The 2014 Nobel Prize in Chemistry awarded three scientists for their outstanding achievements in super-resolution fluorescence microscopy [2].However,in practical biological tissue fluorescence microimaging,most biological tissues have a variety of refractive index inhomogeneities,such as endogenous pigments,blood vessels,and cell clusters [3].These tissues absorb and scatter light,resulting in signal attenuation and aliasing in the propagation path of the final fluorescent signal.The information on sample structure in the image will be degraded,and the image signal-to-noise ratio(SNR)and resolution will be reduced [4].Therefore,deep 3D imaging of biological tissues is a very challenging task.In addition,the quality of the fluorescence signal depends on factors such as temporal resolution,total duration of the experiment,imaging depth,fluorophore density,photobleaching properties,and phototoxicity[5-10].
Currently,there are two main types of solutions to the above limitations: the first approach is optimizing the microscope hardware[11].However,due to the physical limitations of the actual imaging equipment,many factors are difficult to optimize simultaneously.For example,reducing the exposure time can improve the imaging speed,but the image sharpness will be reduced [12].The second approach is to use algorithms that improve the quality of microscopic images to increase the efficiency of extracted information [13].Computational imaging methods can be divided into two categories:traditional image algorithms and deep learning-based image algorithms [14].Conventional image algorithms mainly include transmission and scattering media imaging techniques based on optical memory effects,such as wavefront modulation (WMD) and speckle correlation method(SCM) [15,16].Such methods usually require a combination of hardware devices and algorithms.For example,WMD is an optical modulation technique based on a feedback mechanism.It mainly relies on the spatial light modulator (SLM) to modulate the wavefront of the incident light at the micron level to achieve the effect of making the scattering medium into an imaging lens [17].The imaging principle of the WMD is based on the optical memory effect.The incidence angle of the coherent beam incident on the scattering medium is tilted within a specific range,the distribution of its scattering spots is constant,and the overall translation occurs.Therefore,if SLM can achieve the focus of the transmission scattering medium,the sample near the focus can be reconstructed by changing the angle of the incident wavefront.However,for thicker biological tissues,the effective range of the memory effect is smaller,limiting the field of view for imaging.In addition,the WMD relies on the guide star in the sample to provide light intensity feedback information and then combined with iterative algorithms for phase compensation of the incident light wavefront,so the WMD is challenging to achieve non-contact imaging.At the same time,maintaining the conjugate relationship between the SLM and the sample in position during imaging limits its ability to image in three dimensions.In contrast,deep learning(DL)-based image algorithms are based on a data-driven approach and therefore do not require complex optical hardware.The development of DL has so far gone through three periods of development [18].
1943-1969 (origin phase): Various artificial intelligence concepts were proposed during this period.
1974-2006 (development stage): representative techniques such as backpropagation algorithm,recurrent neural network,convolutional neural network (CNN),and other deep generative architectures were gradually established.
2006-present (explosion phase): DL started to be widely used in various fields of science and technology.
In recent years,the use of DL for biomedical image processing and data analysis has shown a rapid growth trend [19].It can learn the mapping relationship of the inverse scattering process from the scattered image to the clear image by using a large number of scattered-clear image pairs [20].Compared with traditional image reconstruction algorithms,DL has stronger applicability.Once trained on the training set,DL can produce high-quality results under simpler and faster conditions.Meanwhile,DL utilizes neural networks for data-driven statistical inference and does not require numerical modeling of the imaging process or estimation of point spread functions.Based on these advantages,DL has become an effective method to compensatefor theshortcomings offluorescence microscopy.
Reports on the use of DL to improve the performance of fluorescence microscopy date back to 2017.So far,this topic has received much attention from scientists.A series of reviews on this topic have also been published [21-26].They provide a timely summary of the important work on this topic and update the state-of-the-art advances.Therefore,instead of the traditional approach of a repetitive description of this topic,we have adopted bibliometrics in this review to provide a statistically based analysis of this development.In this review,we analyze the development of this topic,including the pattern of development,the contribution of countries/institutions,the direction of the investigation,keyword clustering,and terminology analysis.Of course,the important literature on this topic is also analyzed in detail.Bibliometrics is a quantitative analysis of the literature that provides an understanding of a topic by analyzing the interrelatedness of information in different sections of a paper(e.g.,title,keywords,authors,references).We hope to dig deeper into the information of the development process of this topic through bibliometrics.At the same time,we hope to make reasonable predictions regarding the direction of this topic based on the available information.
Materials and methods
Two bibliometrics software have been used in this systematic literature review.The first is CiteSpace,developed by Dr.Chaomei Chen,a professor at the Drexel University School of Information Science and Technology [27-30].CiteSpace 6.1R2 was used to calculate and analyze all documents.In addition,the bibliometrix package of R was used to perform some additional scientometric analysis[31].
Core collection on the Web of Science has been selected as a database to assure the integrity and academic quality of the studied material.“‘deep learning’‘fluorescence microscopy’”has been used as a“Topic”.The retrieval period was indefinite,and the date of retrieval was July 2022.158 articles (including 105 articles,12 reviews,41 proceeding papers,2 early access,1 correction,and 1 data paper)were retrieved.Articles,proceeding papers,early access,and data paper were selected among them,remaining 145 publications.We then conducted further manual screening of these papers by title and abstract.Some of these references were excluded because although they included “deep learning” and “fluorescence microscopy” in the retrieved content,they did not investigate this topic.At the same time,some reviews were recorded in WOS as research articles due to information errors.In total,138 articles remained in the dataset.Figure 1 shows the flow chart of data acquisition.
Developments in the research field
Literature development trends

Figure 1 Flowchart of search and refining on fluorescence microscopy with DL in Web of Science,through the different phases of the systematic review.DL,deep learning.

Figure 2 Annual publications from 2017 to 2022 searched in the WOS about fluorescence microscopy with DL.DL,deep learning;WOS,Web of Science.
The number and distribution of papers published can be used to understand the evolution of a topic.DL and fluorescence microscopy were combined for the first time in 2017.Rivenson et al.applied DL to wide-field microscopy image conversion [32].They trained the with CNN,resulting in significant improvements in the spatial resolution performance of wide-field microscopy over a large field of view and depth of field.Figure 2 shows the annual number of articles published on this topic from 2017 to date.As can be seen,this topic received much attention immediately after the first article was published in 2017.The annual publication exceeded 10 articles in both 2018 and 2019.This topic drew its first apex in 2020,with 41 publications.During these four years,investigations on this topic showed explosive growth,but this trend did not last.The annual publication number on this topic showed a slight decrease for the first time in 2021,with 39 articles published.As of July 2022,the annual number of articles on this topic was 25.Due to the regular cycle of academic paper publication,we guess the annual number of publications this year should be the same as the previous two years,and there will not be a significant increase or decrease.As a result,this topic has entered a plateau in recent years.Based on the pattern of technology migration,it is possible that the explosion of annual publications on this topic in the first four years was the result of different scientists trying to migrate already mature algorithms to fluorescence microscopy applications.In this process,the challenges of DL in the application of fluorescence microscopy were gradually observed.These challenges cannot be overcome by simply applying the general algorithms that have been established,so the research progresses to a plateau.This topic received much attention as soon as it appeared in academia,with an annual growth rate of 90.37% of published papers.At the same time,these published papers have received significant attention,with an average citation per paper of 10.93.
Journals,cited journals,and research subjects
The 138 papers retrieved were published in a total of 91 different journals,representing the scopes of journals in which this topic fits into different areas.This phenomenon is generally more common in cross-disciplinary academic topics.Figure 3 shows the 9 journals with the highest number of publications.Among them,Biomedical Optics Express published the most papers.Optics Express ranked second with a total of 7 papers.As this topic deals with technology in the field of computing,the conference papers also make an important contribution to this topic.4 and 3 conference papers related to this topic were presented at ISBI 2020 and ISBI 2021,respectively.From the types of journals,it can be concluded that optical,medical,bioinformatics and multidisciplinary journals are the ones that publish the most on this topic.
In addition to the journals in which the papers are published,local cited journals are also essential information to understand what areas the content of the topic is related to.Figure 4 shows the top 20 local cited journals on fluorescence microscopy with DL.Among them,the paper in Nature Methods is widely cited on this topic.Since we retrieved only 138 papers,the 475 citations represent a series of papers published in Nature Methods that greatly impacted the field.Among them,the paper published in 2018 by Weigert et al.received the most extensive citations [33].They recover the quality of fluorescence images with a 60-fold difference in photon number for networks trained with low SNR images as input and corresponding high SNR images as the gold standard.This study shows that DL allows for rapid sampling of light-sensitive and highly dynamic fluorescent samples.The super-resolution supported by DL in different fluorescence microscopy modes was presented by Wang et al.[34].The use of DL to recover fluorescence information can be understood as training a model for image noise reduction or weak signal enhancement.They train the network with low-resolution confocal data as input and the corresponding stimulated emission depletion(STED) data as the gold standard.The resulting model helps researchers to increase the resolution of data taken by confocal to a level close to that of STED without the need for super-resolution imaging equipment.
Since the content of this topic is closely related to the progress of DL-assisted image recognition,a series of important IEEE conferences are also included in Figure 4.In addition,some journals in Figure 3 do not appear in Figure 4,such as Biomedical Optics Express and Bioinformatics.This further represents that the underlying methodology of this topic is based on other related topics.In addition,since the topic has only undergone 5 years of development,it has not yet been widely used in practical applications.Therefore,some journals have published papers on this topic but have not been widely cited.Although the journals in Figure 3 and Figure 4 do not overlap exactly,they both focus on several areas,including medical,optical,and multidisciplinary journals.
To learn more about the latest developments in fluorescence microscopy with DL,we counted the journals that were first published on the topic in 2022,including Acta Neuropathological Communications,Chinese Physics B,Genes,Information,Neural Computing &Applications,Neural Networks,New Phytologist,Open Biology,Optics and Lasers in Engineering,Photoacoustics.As can be seen,the latest journals on this topic have specific application directions,including botany,neuropathology,and biology,in addition to optics and computer science.We suspect that DL-assisted fluorescence microscopy has begun to be used to address some of the challenges once faced in experimental and clinical research.
Geographic and author distribution
Figure 5 shows the top 16 countries (corresponding author's country)with the most publications.The USA contributed the most papers on this topic,participating in 38 papers.China was second only to the USA regarding the number of papers published,participating in a total of 34 papers.However,China prefers international collaboration,with 6 papers belonging to multiple country publications.Most of the top 10 countries,except for Korea and France,have formed international cooperation networks.
In addition to the number of papers a country publishes,the number of citations it receives for its papers also reflects its contribution to the subject.Table 1 shows the top 10 countries in the number of citations and their average article citations.The papers published by the USA were the most widely cited,with over 600 citations.The second-ranked country is Germany,with a total of nearly 400 citations.Meanwhile,Germany has the highest average article citations,with 23.18.In contrast,China has not attracted many citations,although it has published several papers on this topic.Although the ranking order in Figure 5 and Table 1 do not precisely match,the countries in Table 1 all appear in Figure 5.This means that the countries most involved in this topic are also the leading promoters of this topic.

Figure 3 Top 9 journals that published articles on the fluorescence microscopy with DL.DL,deep learning.

Figure 4 Top 20 located cited journals on the fluorescence microscopy with DL.DL,deep learning.

Figure 5 Top 16 countries of paper publications to the topic of fluorescence microscopy with DL.DL,deep learning.

Figure 6 Time-zone view of research categories for fluorescence microscopy with DL.DL,deep learning.
Figure 6 illustrates the evolution of fluorescence microscopy with DL among different categories.Since only one paper was published on this topic in 2017,the category only covers Biochemical Research Methods and Microscopy.Most of the categories related to this topic have been covered in the subsequent 2018,mainly including biology,medicine,and computer science.In 2019,Engineering(Electronical &Electronic),Physics (Applied),Cell Biology,and Pathology were included.Starting in 2020,the field of materials science and chemistry-related fields also enters this topic.The evolution of category partially confirms our previous suspicions.DL-assisted fluorescence microscopy began to be used in different image processing and recognition after undergoing algorithmic adaptation.
Geographic and author distribution
Figure 5 shows the top 16 countries (corresponding author's country)with the most publications.The USA contributed the most papers on this topic,participating in 38 papers.China was second only to the USA regarding the number of papers published,participating in a total of 34 papers.However,China prefers international collaboration,with 6 papers belonging to multiple country publications.Most of the top 10 countries,except for Korea and France,have formed international cooperation network.
In addition to the number of papers a country publishes,the number of citations it receives for its papers also reflects its contribution to the subject.Table 1 shows the top 10 countries in the number of citations and their average article citations.The papers published by the USA were the most widely cited,with over 600 citations.The second-ranked country is Germany,with a total of nearly 400 citations.Meanwhile,Germany has the highest average article citations,with 23.18.In contrast,China has not attracted many citations,although it has published several papers on this topic.Although the ranking order in Figure 6 and Table 1 do not precisely match,the countries in Table 1 all appear in Figure 6.This means that the countries most involved in this topic are also the leading promoters of this topic.
Figure S1 illustrates the cooperative relationship between different countries.First,this topic has not attracted the attention of the global academic community,and the Middle East region and the South American region are not involved in this topic.International cooperation is mainly carried out with the USA/China,USA/Japan,and European countries.Table 2 shows the 9 affiliations with the highest number of publications on fluorescence microscopy with DL.UCLA participated in the publication of 37 papers on this topic,presenting a clear advantage in terms of the total number.Huazhong University of Science and Technology came in second place with 22 publications.UC San Francisco,Heidelberg University,and The University of Tokyo were all involved in at least 10 papers.The affiliations in Table 2 include USA,China,Germany,Japan and the UK.These affiliations are the top-ranked countries in Figure 6,except for Japan.This means that although Japanese scientists have not published many papers as corresponding authors,they have been extensively involved in the investigation of this topic.

Table 1 Top 10 countries of total citations on the topic of fluorescence microscopy with DL with the average article citations
Figure 7 shows the collaboration of the different affiliations on this topic.It can be seen that fluorescence microscopy with DL has gradually formed two collaborative networks in the 5 years of development.Uppsala University and the University of Zurich led the largest collaborative network.The network includes universities and research institutes in Sweden,Switzerland,Finland,UK,Germany,and the Czech Republic.Another cooperation network is led by Huazhong University of Science and Technology and the Chinese Academy of Sciences.This cooperation network mainly includes Chinese and German universities and research institutes.UCLA,which has the most publications in Table 2,also collaborates with The Hebrew University of Jerusalem and The Ohio State University but has yet to form a complex collaboration network.
Figure 8 shows the top 10 authors who are active in this topic.An analysis of the published papers of these authors reveals that they belong to three different groups.Peng Fei has published 7 papers in less than 3 years,although it only became involved in investigating this topic in 2020 [35-41].He proposed several DL algorithms with co-workers to improve the imaging results.Different types of fluorescence microscopy assisted by DL have shown advanced performance,such as recording the neural activity during forwarding and backward crawling of 1st instar larvae;investigating the complex spatial relationships and interactions between the endoplasmic reticulum and mitochondria throughout living cells [35,37].Karl Rohr,Ralf Bartenschlager,Roman Spilger and Ji-Young Lee have contributed 7 papers in this topic as well [42-48].They and co-workers focus on the performance of different DL algorithm improvements for particle tracking in the fluorescence image.Aydogan Ozcan,Laurent A.Bentolila,Yilin Luo,Hongda Wang,and Aydogan Ozcan have not published as many papers as the previous authors,but they received much attention for their publications in 2019.One of their works has already been described in section 3.2[34].In another paper published in Nature Method,they used trained deep neural networks to refocus a two-dimensional fluorescence image onto user-defined three-dimensional sur-faces within the sample [49].This technique,which they named Deep-Z,can increase the depth of the field of view by a factor of 20.
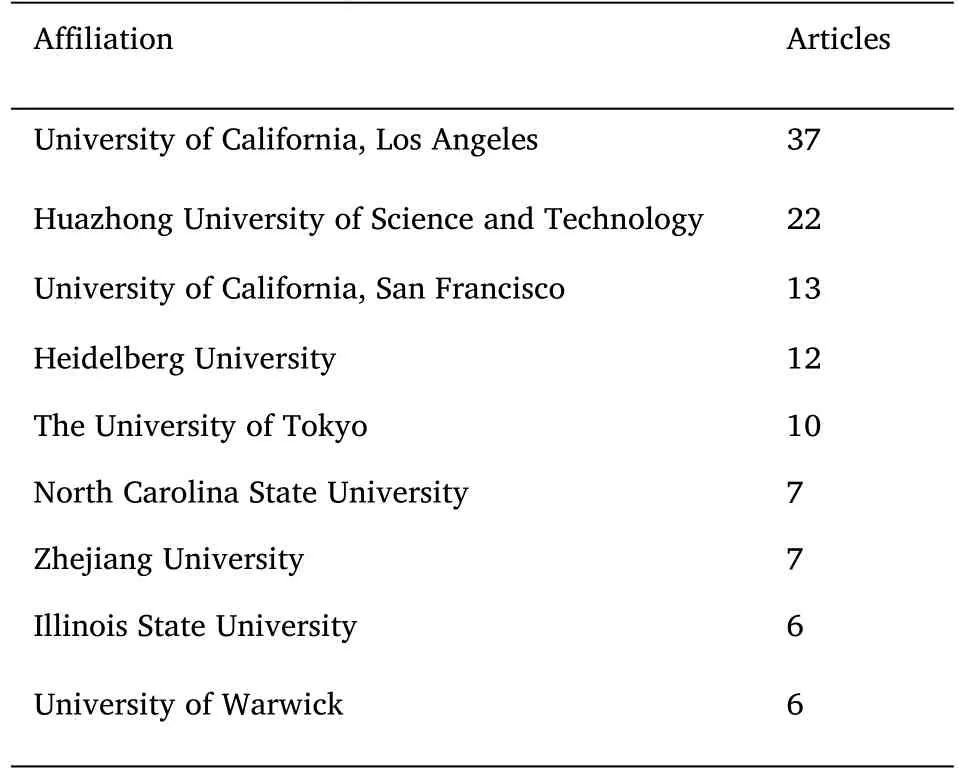
Table 2 Top 9 affiliations of paper publications to the topic of fluorescence microscopy with DL
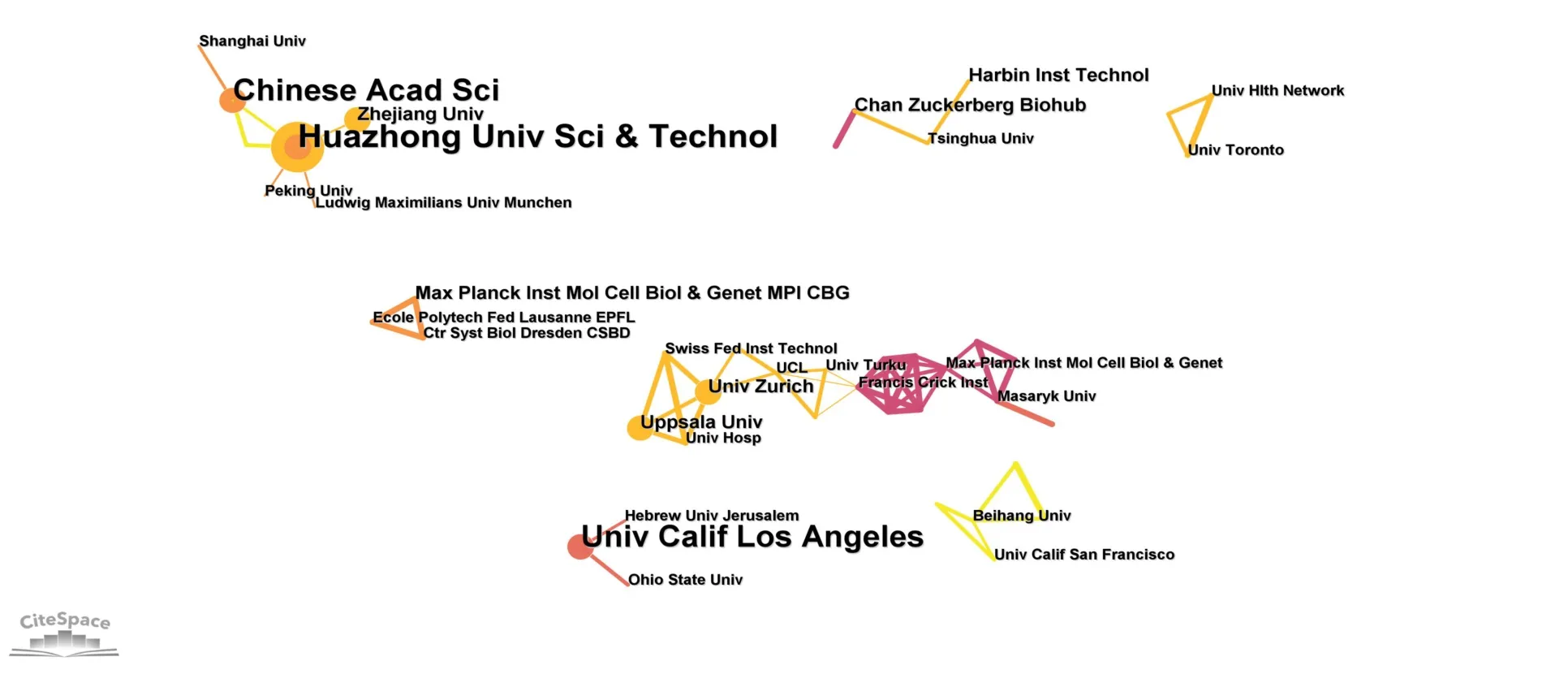
Figure 7 Affiliations cooperation network of published papers for fluorescence microscopy with DL.DL,deep learning.

Figure 8 Top 10 author production over time in the topic of fluorescence microscopy with DL.DL,deep learning.

Figure 9 Top 12 most frequent words in author’s keywords in the topic of fluorescence microscopy with DL.DL,deep learning.
A Sankey diagram was created to show more clearly the relationships and contributions made by authors,institutions,and countries on this topic (Figure S2).
Keyword,term analysis and evolution of the field
Keywords,term analysis
The statistical analysis of the words in different parts of the paper gives an idea of the different research directions on a topic.Figure 9 shows the most frequent words calculated from the author's keyword(keywords provided by the author in the paper).Not surprisingly,deep learning and fluorescence microscopy were the most frequently occurring keywords,appearing 46 and 31 times,respectively.Convolutional neural networks were the most frequently appearing keyword subsequently.As a feed-forward neural network,CNN is widely used in image processing.CNN essentially extracts the local information of the image to be processed by sliding the convolutional kernel over the image to be processed.The image processed by the convolutional kernel is called the feature map.The feature map is selected and retained by subsequent pooling layers and activation functions.Finally,the image's local and global features are extracted by successive convolution operations to complete the noise reduction of the whole image.Since winning the ISBI Cell Tracking Competition in 2015,U-Net has become one of the most popular CNNs available(Figure 10) [50].U-Net is named after its “U” shaped network structure and can be divided into two processes: downsampling and upsampling.Among them,convolutional and pooling layers mainly implement the downsampling process.It enhances the robustness of the model against noise interference while extracting image features.It also reduces the risk of overfitting,reduces the number of operations,and increases the size of the perceptual field.The upsampling process is mainly implemented by deconvolution,which can reduce and decode the abstract features into the dimensions of the input image and obtain the final processing result.In addition,compared with traditional convolutional networks,U-Net uses layer-hopping connections,thus solving the problem that the original input information is gradually lost as the network depth increases.This makes it possible to get good training results even on small sample training datasets.In particular,the excellent performance in the cell segmentation project demonstrates the capability of the U-Net network in extracting image features and correctly decoding the features to recover the image according to the task requirements[51].
The fourth-ranked keyword is segmentation.The introduction of DL into the field of image processing has given more accurate semantic information to the segmented region,while the image segmentation problem has made a breakthrough.The emergence of image segmentation methods based on deep learning,such as full convolutional network,pyramid scene parsing network,DeepLab,Mask R CNN,etc.,has led to increased accuracy of segmentation and a more intelligent process of segmentation [52-55].The main challenge faced in fluorescence micrographs is the segmentation of cells.Cell segmentation refers to the automatic computerized differentiation of single cells from the background and other cells in 2D or 3D microscopic images for subsequent processing,such as single-cell feature extraction,cell typing,or cell tracking.The previously mentioned U-Net framework became a common structure for processing biological images in the medical field,and many related and improved frameworks were derived [36,37].This framework's publication changed the cell segmentation field,and the construction of neural network-based segmentation models has since become mainstream [56,57].The U-Net,StarDist,and YOLO algorithms have all been used for image segmentation of fluorescence microscopy images [58-60].U-Net and subsequent DL network-based cell segmentation methods transform the cell segmentation problem into a pixel-wise classification problem.Typically,each pixel location on a cell image can be classified into one of three categories: intracellular,cell edge,and background.Therefore,neural networks are usually trained and supervised by labeled images.However,annotating an image usually takes more than half an hour,resulting in a high acquisition cost and limited training set data.In 2017,Sadanandan et al.automatically developed a method to acquire training sets by fluorescence imaging [61].By fluorescently labeling cell structures and simple image processing,it is possible to automate the acquisition of high-quality data annotation sets and train neural networks with bright-field images and annotated data.In recent years,the field of DL-based cell segmentation has also been exploring other possible frameworks.For example,Wang et al.published a novel cell segmentation framework combining two DL networks and traditional machine vision processing algorithms [62].In this framework (Figure 11)[62],the neural network does not directly classify each pixel point of the image but learns the similarity between pixel points and gives an Eulerian distance representation of the cell pixel similarity.A second neural network is then used to obtain the cell location in the form of target detection,and finally,the segmentation results are obtained using traditional computer vision algorithms.This framework can be an excellent solution to the problem of under-segmentation or over-segmentation in the DL segmentation framework based on pixel classification.

Figure 10 U-Net architecture.

Figure 11 Combines CNN and watershed for single cell segmentation.CNN,convolutional neural network.
Image reconstruction (ranked 7th in Figure 9) is also an important task for DL in fluorescence microscopy image processing.Common image reconstruction algorithms include statistical super-resolution algorithms based on the physical properties of fluorescent molecules,deconvolution algorithms based on the corresponding imaging models,surface projection algorithms based on morphological models,and data-driven DL algorithms [63,64,55,56].All the above algorithms can cleverly bypass the physical limitations and overcome the shortcomings of fluorescence microscopy to recover biological information better.Compared to other algorithms,DL can more fullyexploit the prior knowledge of existing data and recover more information for a specific task [65].For example,in confocal microscopy,DL reconstructs the acquired low-scan density image into a high-scan density image,which can speed up the scanning process of low-scan density confocal microscopy imaging while obtaining the same quality high-scan density image.Li et al.proposed a back-projection generative adversarial network,which solved the slow imaging speed of confocal fluorescence microscopy [66].back-projection generative adversarial network uses a generative adversarial network (GAN,the 9th ranked keyword in Figure 9)-based approach to obtain similar quality to high-resolution confocal scans with a 64-fold increase in imaging speed.
In Figure 9,two types of fluorescence microscopy become high-frequency keywords: light-sheet fluorescence microscopy and structured illumination microscopy.Light-sheet fluorescence microscopy is a novel three-dimensional imaging fluorescence microscopy technique.A cylindrical mirror focuses its illumination light in a single dimension to form a thin sheet of light parallel to the imaging plane.Only the sample to be imaged on the focal plane will be illuminated,achieving the effect of optical slicing.By scanning the sample,the light sheet microscope allows for faster 3D imaging of large samples.Further rotation of the sample gives multi-angle imaging results of the sample.Image fusion algorithms process these multi-view images to obtain isotropic 3D images in the XYZ direction.However,light-sheet fluorescence microscopy has a low axial sampling rate when performing fast volume imaging.This anisotropy affects the accurate measurement.Weigert et al.proposed a content-aware image restoration network,which successfully recovered low signal-to-noise ratio 3D projection images of fruit fly fluorescence films to overcome this shortcoming[33].Structured light illumination microscopy is used to illuminate the sample with a modulated beam containing stripes.It obtains multiple imaging results by changing the direction and phase of the illumination pattern and then relies on image reconstruction algorithms to obtain super-resolution images.A structured light illumination microscopy system has twice the spatial resolution of ordinary wide-field microscopes.It can image living cells for long periods at low illumination levels,making them the most widely used super-resolution microscopy system for in vivo bioimaging.However,a structured light illumination microscopy system requires complex and expensive optical hardware for modulated illumination.It requires multiple measurements to reconstruct a super-resolution image,which is also limited by artifacts.Wang et al.proposed a GAN-based framework for cross-modal hyper-discrimination imaging[34].The framework can convert diffraction-limited images to super-resolution images,better overcome structured illumination microscopy’s shortcomings that require expensive optical hardware for modulated illumination and enable the popularization of super-resolution microscopy.Jin et al.proposed a method to improve the imaging effect of structured illumination microscopy using U-Net[67].They used this method to reduce the number of original image frames required for structured illumination microscopy reconstruction of super-resolution images from 9 to 3.Ling et al.proposed a CNN-based fast light-sheet fluorescence microscopy method,which reduced the required number of frames of the original image by 4/5[68].
To further mine the information on word occurrence frequency,we counted the word frequency of keywords plus (keywords automatically added by WOS) and the abstract section,and the results are shown in Figure S2 and S3.Since the system automatically captures the words in these two sections,they do not always accurately convey the focus of the paper.Some of these high-frequency words have a high match with the results in Figure 9,such as particle tracking,neural network,etc.In addition,some high-frequency words are additional elements in the keywords in Figure 9.For example,cell nuclei in Figure S4 focus on cell segmentation that needs to be identified.The signal-to-noise ratio is an important index of signal reconstruction quality in image noise reduction,and there is a positive correlation between its value and reconstruction results.Depth-of-field represents the extent of the image’s field of view.The Deep-Z model mentioned in Section 3.3 allows the calculation of dynamic information of a live sample in 3D space based on time-series fluorescence images acquired on a single focal plane.This technique increases the depth of field by a factor of 20 without axial scanning.
Cluster analysis
Clustering analysis of keywords allows the analysis of different directions under one topic.Figure 12 shows the clustering analysis of keywords using CiteSpace.The different clusters in this topic do not show significant overlap,representing a large variability between the clusters.Table 3 shows the size,Silhouette value,representative keywords,and representative papers of different clusters.The following is a brief description of each cluster.
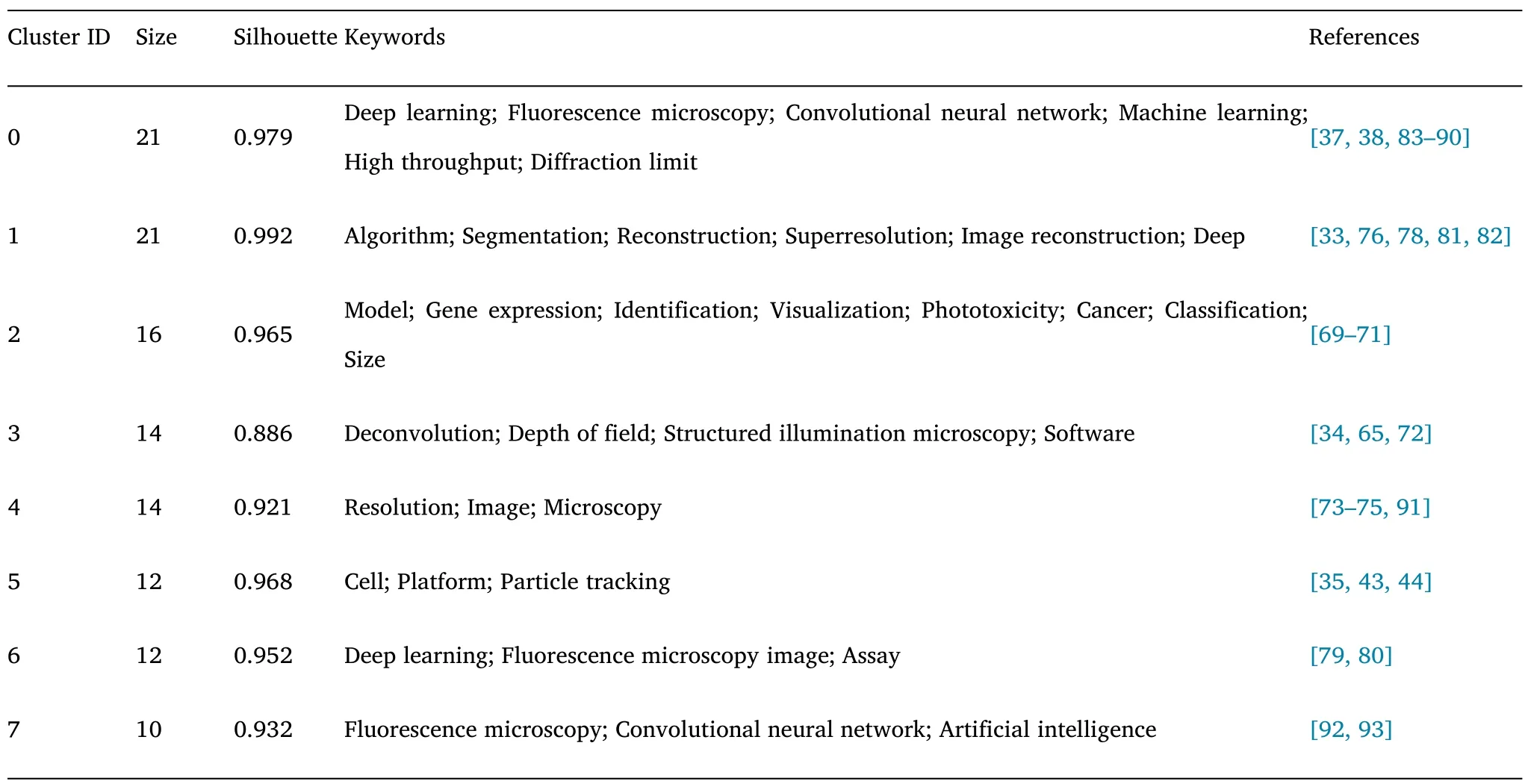
Table 3 Knowledge clusters in the field of fluorescence microscopy with DL on keyword co-occurrences for each cluster
#0 (Deep Learning) This cluster contains the largest number of papers.This series of papers is fundamental research on this topic.They build themain algorithms and techniques for this topic.
#1 (Localization) This cluster mainly gathered attempts to achieve efficient localization in fluorescence microscopy images.Jiang et al.proposed solving the interference of plant autofluorescence to achieve efficient image analysis [69].Bao et al.proposed a multi-modal structured embedding technique that can integrate multi-modal data to gain insight into cells’ state,function,and organization in complex biological tissue [70].Tahir et al.proposed an end-to-end Protein localization CNN specifically for protein localization analysis [71].
#2 (Wide-field image) The papers in this cluster focus on how to acquire wide-field images with high resolution.Wang et al.convert diffraction-limited input images to super-resolution images based on GAN,which results in improved resolution for acquiring wide-field images with low numerical aperture objectives [34].Luo et al.proposed a Deep-R autofocus method for fast microscopic imaging in large sample areas [72].
#3 (Morphology analysis) The morphological analysis of different cells and organelles is one of the tasks that fluorescence microscopy can accomplish.The papers in this cluster contain the efforts of different research groups in this direction.Fischer et al.developed a MitoSegNet tool to analyze mitochondrial morphology [73].Lee et al.proposed a method specifically for the kinetic analysis of immune synapses [74].Zhang et al.proposed a technique for analyzing single-cell morphology in bacterial biofilm [75].
#4 (Particle tracking) This cluster mainly brings improvements in automatic particle tracking methods.For example,Spilger et al.proposed recurrent neural network architecture for this purpose in each two papers [43,44].
#5 (Illumination control) The representative papers contained in this cluster can be divided into two different directions.The first sub-topic is the determination of optimal acquisition parameters through illumination control and DL [76,77].The second sub-theme is to provide more efficient content-aware image restoration effects[33-78].
#6(Dynamic imaging)The two representative papers in this cluster deal with dynamic fluorescence imaging.Tahk et al.investigated the M4 muscarinic acetylcholine receptor imaging with fluorescent ligands [79].By fluorescence microscopy,they developed a technique to characterize the affinity between living CHO-K1-hM4R cells and ligands.Akerberg et al.developed an analytical method to extract volumetric parameters of cardiac function from dynamic light-sheet fluorescence microscopy images of zebrafish embryonic hearts [80].
#7(Cell)Two of the works included in this cluster are related to cell segmentation and recognition.Raza et al.proposed an improved CNN-DL architecture for segmenting objects in microscopy images[81].Sullivan et al.created a mini-game to engage players in cell classification [82].Combining player annotations and DL,they developed an automated tool for localizing cell annotations.
From the regional distribution of different clusters,it can be seen that the research of deep learning in fluorescence microscopy mainly focuses on the innovation and establishment of methodology.Thus,the clusters in the central area focus on technical details and overlap with each other.The clusters on the periphery are mainly for the specific application of deep learning for fluorescence microscopy.
Conclusions
Fluorescence microscopy is widely used in life science because of its non-destructive,non-contact,high specificity,and high sensitivity.However,because of the optical diffraction limit,conventional fluorescence microscopy cannot observe fine structures in cells below approximately half a wavelength.These limitations have led researchers to look to data-driven DL.As a rapidly developing new technology,DL has received increasing attention in microscopic image processing,mainly focusing on tasks such as image segmentation and classification,target recognition,and tracking.Combining DL to build intelligent microscopy-assisted software or intelligent microscopes will become a mainstream trend in imaging.In this bibliometric-based review,we not only interpret the important papers on this topic but also analyze the development of the whole topic from a statistical perspective.The following results were obtained based on bibliometric analysis:
This topic has experienced just under six years and exploded during the first three years.The number of papers published in this field entered a plateau in the last three years.Based on the analysis of paper institutions and authors,this topic has attracted a stable research group,so it will not be a flash-in-the-pan topic.
Papers on this topic are mainly published in optical,medical,bioinformatics,and multidisciplinary journals.In addition,papers published in Nature Methods related to DL-assisted image processing have been very influential in developing this topic.A series of conference papers on image processing organized by IEEE also play an important role in this topic.
Although this emerging topic has attracted much attention,the researchers interested in it show a clear regional character.The Middle East and South American regions are not involved in this topic.Meanwhile,although established,international cooperation does not present a very complex network.International cooperation is mainly carried out with the USA/China,USA/Japan,and European countries.
This topic already has several established research groups,many of which have been making new contributions to the topic every year since 2018 without interruption.This is a very important factor for the long-term development of a topic.At the same time,they are likely to develop complex collaborative networks that will further drive the topic to address the remaining challenges.
According to the keyword analysis,the application of DL in fluorescence microscopy imaging can be divided into three main directions.The first direction is accelerating the reconstruction speed of images;the second is accomplishing cross-system imaging by using different optical imaging systems;the third is using two-dimensional data to reconstruct three-dimensional information.
Perspectives and challenges
In addition to the above conclusions,the bibliometric analysis can provide perspectives on future directions through the changing information on a topic.In combination with a series and reviews and perspective articles,we also provide some perspectives on this topic:
The simple migration of methodologies of DL in other fields is not perfectly adapted to the samples faced by fluorescence microscopy.Therefore,the targeted optimization of different DL frameworks based on the optical properties of fluorescence microscopy and sample characteristics is a shortcut.
Training DL networks requires high computational costs and often requires researchers with coding and computational expertise.However,for many biomedical researchers,the lack of relevant knowledge hinders the application of the technology.Therefore,the availability of cloud-based trained DL models and manuals equipped with recommendations for their operation can significantly increase the potential for application.However,there is a risk of prediction errors if the models are used on new data with insufficient similarity to the training data.
Current computational imaging approaches based on DL can use feature information extracted from many samples to model the actual imaging process,reducing the need for accurate modeling.However,the essence of DL is to summarize the statistical properties that are“common” to a large number of training samples.However,for some specific applications,such as medical testing and lesion screening,users tend to focus more on the specificity of the samples.The accuracy of DL for reconstructing “rare samples” (which are significantly different from the training data set) is not ideal.
All DL-related fields face a common problem: our understanding of DL in principle is still insufficient.Therefore,the underlying breakthrough of DL is believed to achieve a new revolution in fluorescence microscopy imaging analysis.
- Medical Data Mining的其它文章
- Highly expression of MYBL2 is correlated with a poor prognosis and immune infiltration in clear cell renal carcinoma
- Bibliometric analysis of literature review of osteoarthritis in the Cochrane database
- Simulation analysis and pharmacokinetic evaluation of atractylide I for the treatment of diabetic nephropathy
- Feature selection and machine learning approach for carotid atherosclerosis in asymptomatic adults
- Investigating public perceptions regarding the Long COVID on Twitter using sentiment analysis and topic modeling
- A novel prognosis signature based on two cuprotosis-related genes for kidney renal clear cell carcinoma

