OsSPL10 controls trichome development by interacting with OsWOX3B at both transcription and protein levels in rice (Oryza sativa L.)
Qioling Lio, Xinle Cheng, Tong Ln, Xiokun Guo, Zilong Su, Xioxio An, Yli Zheng,Hito Cui, Weiren Wu,b,*, To Ln,b,c,*
aKeyLaboratoryof Genetics, Breeding and MultipleUtilizationof Crops, Ministry of Education, FujianAgriculture and Forestry University, Fuzhou 350002, Fujian,China
bFujian ProvincialKey LaboratoryofCrop BreedingbyDesign,Fujian Agriculture and Forestry University, Fuzhou 350002, Fujian,China
cKeyLaboratoryof Biological Breeding for Fujian andTaiwan Crops,Ministry of Agriculture and RuralAffairs,Fujian Agriculture and ForestryUniversity,Fuzhou350002,Fujian,Chi na
Keywords:Rice Trichome OsSPL10 OsWOX3B HL6 Interaction
ABSTRACT Plant trichomes are a specialized cellular tissue that functions in resistance to biotic and abiotic stresses.In rice, three transcription-factor genes: OsWOX3B, HL6, and OsSPL10, have been found to control trichome development.Although studies have shown interactions between the three genes, their full relationship in trichome development is unclear.We found that the expression levels of OsWOX3B and HL6 were both reduced in OsSPL10-knockout plants but increased in OsSPL10-overexpression plants,suggesting that OsSPL10 positively regulates their expression.Physical interaction between OsSPL10 and OsWOX3B was found both in vivo and in vitro and attenuated their abilities to bind to the promoter of HL6 to activate its transcription.This mechanism may regulate trichome length by adjusting the expression of HL6.A rice gene network regulating trichome development is proposed.
1.Introduction
Trichomes (epidermal hairs) are specialized plant structures that originate from aboveground epidermal tissues and develop into hairlike projections extending from epidermal surfaces during growth, differentiation, or cell division [1].Trichomes can protect plants from insect herbivore and pathogen damage, reduce the influence of strong light, and regulate transpiration [2].Trichome development has been extensively studied, not only for its importance but also as a mechanistic model for epidermal cell differentiation and cell morphogenesis.
As a model plant,Arabidopsis has been the subject of trichomedevelopment studies.The R2R3 MYB transcription factor gene GLABRA1 (GL1) was the first cloned gene influencing trichome development in Arabidopsis [3].The GL1 protein combines with the bHLH transcription factor GLABRA3 (GL3) [4] and the WD40 repeat protein TRANSPARENT TESTA GLABRA1 (TTG1) [5] to form a heterologous complex named MBW, which activates the expression of the homeodomain gene GLABRA2 (GL2) to promote trichome development [6–8].
In rice, four genes critical for trichome development have been cloned: OsWOX3B (also named DEP, NUDA/GL-1 or GLR1), HL6(OsPLT2), OsSPL10 (SST), and LHL1.OsWOX3B encodes a WUSCHEL-like homeobox (WOX) transcription factor.The WOX family is a plant-specific transcription factor family with 13 members in rice.They function in meristem establishment,lateral organ development, floral organ formation, and plant stem and root apical meristem development [9].Loss of OsWOX3B function causes absence of trichomes on leaves and glumes, suggesting that OsWOX3B is required for trichome initiation and elongation [10–12].OsSPL10 encodes an SQUAMOSA PROMOTER BINDING PROTEIN (SBP) box transcription factor.The SPL gene family members encode transcription factors unique to green plants,which contain a highly conserved SBP domain[13].In rice,18 members of the SPL family have been identified, controlling a wide range of processes underlying plant growth and development [14,15].Knockout of OsSPL10 results in glabrous leaves and glumes,whereas its overexpression increases leaf trichome density [16].Thus, OsSPL10, like OsWOX3B, is necessary for trichome initiation and elongation.HL6 encodes an AP2 (APETALA2)/ERF (ethylene-responsive factor)-type transcription factor.Such transcription factors represent a large family of proteins characterized by the presence of an AP2 domain.They regulate diverse processes of plant development and stress responses [17,18].HL6 is necessary for trichome elongation.It determines trichome length by regulating the expression of OsYUCCA5 and other genes responsible for auxin synthesis.Low HL6 expression leads to small trichomes visible only by light microscopy [19].Unlike the above three genes, which all encode transcription factors, the newly identified [20] gene LHL1 encodes a plant-specific, highly conserved nuclear protein that contains a RibonucleaseH-like (RNaseH-like) domain.LHL1 also positively regulates leaf trichome development and is also probably associated with an auxin pathway [20].
The relationships among the four genes in control of trichome development have been partially clarified.OsWOX3B activates the expression of HL6 by binding to the promoter of HL6, and can also combine with HL6 into a complex that increases the binding ability of HL6 to the promoter of the target gene OsYUCCA5 [19].OsSPL10 binds to the promoter of HL6 to activate its expression[21].LHL1 positively regulates the expression of both OsWOX3B and HL6 [20].These findings indicate the connections among the four rice genes, suggesting that they form a regulatory network for trichome development.
In this investigation,a combination of biochemical and molecular biological techniques was employed to explore the transcriptional regulation mechanisms involving OsSPL10, OsWOX3B, and HL6.Moreover, the study also examined the protein interaction between OsSPL10 and OsWOX3B, along with the subsequent impact of this interaction on the transcription of HL6.By utilizing the findings derived from these experiments, the primary goal was to gain a deeper understanding of the intricate relationship existing among OsSPL10, OsWOX3B, and HL6, specifically in their role as regulators of rice trichome development.
2.Materials and methods
2.1.Plant materials
Three rice mutants, Osspl10, Oswox3b, and hl6, were used.Osspl10 was generated from the japonica rice cultivar Zhonghua 11(ZH11) by CRISPR/Cas9 editing, in which bases A and T were inserted into their respective target sites in the first exon of OsSPL10, resulting in premature termination of translation(Fig.1A).Osspl10 displayed glabrous leaves and glumes as reported[16].Oswox3b was obtained from the indica rice cultivar R401 by CRISPR/Cas9 editing, by which base 341 was changed from T to C and bases 395–397 were deleted,resulting in respectively a change from valine to alanine and a loss of serine in the first exon of OsWOX3B (Fig.1A).Oswox3b also displayed glabrous leaves and glumes as reported [22].hl6 was generated from the japonica rice cultivar ZH11 by T-DNA insertion in the sixth intron of HL6(Fig.1A)and showed very small trichomes on leaves[19].OsSPL10-overexpression lines generated from ZH11 and showing higher density of macrohairs on leaves [16] were also used.
2.2.RNA isolation and qRT–PCR analysis
After pregermination at 26 °C for 2 d, rice seeds were sown in paddy soil in plastic trays and grown at 25±2°C under a photoperiod of 14 h light/10 h dark.The shoots of 14-day-old seedlings were collected,with at least 6 plants pooled for each sample.Total RNA was extracted using a Trizol Kit (Invitrogen, Carlsbad, CA,USA) according to the manufacturer’s instructions.The RNA was treated with DNase I and used for first-strand cDNA synthesis with MLV reverse transcriptase (Invitrogen).qRT-PCR was performed using SYBR Premix Ex TaqTM(Tli RNaseH Plus;Takara)on a Prism 7500 96 Real-time PCR System (ABI), and the rice gene Actin was used as internal control.The primers used are listed in Table S1.Three biological replicates were used.Relative quantification was performed using the 2-ΔΔCTmethod [23].
2.3.Yeast two-hybrid (Y2H) assay
The full-length coding region of OsSPL10 was introduced into the yeast expression vector pGBKT7 (Clontech), yielding the construct pGBKT7-SPL10.The full-length CDS regions of OsWOX3B and HL6 were introduced into the yeast expression vector pGADT7(Clontech), yielding the constructs pGADT7-WOX3B and pGADT7-PLT2, respectively.The truncated coding regions of OsSPL10 and OsWOX3B were amplified and inserted into vector pGBKT7 or pGADT7, yielding the following constructs: pGBKT7-SPL10-N(amino acids 1–179), pGBKT7-SBP (amino acids 180–258),pGBKT7-SPL-C (amino acids 259–426), pGADT7-HD (amino acids 1–109) and pGADT7-WUS (amino acids 211–284).The primers used are listed in Table S1.The fusion constructs were used to co-transform the Y2HGold (Weidi, Shanghai, China) yeast strain.The transformed strains were grown on SD/-Leu/-Trp medium at 30 °C for 3 d, and protein interactions were tested on SD/-Leu/-Trp/-His/-Ade selective medium.
2.4.Bimolecular fluorescence complementation (BiFC) assay
The coding sequences of OsSPL10 and OsWOX3B were amplified and inserted into vectors pCAMBIA1300S-YN and pCAMBIA2300SYC, respectively (primers are listed in Table S1).OsH2B-mCherry was used as a nuclear localization marker.Rice protoplasts were prepared using the shoots from 14-day-old seedlings, and various plasmid combinations were introduced into protoplasts and incubated at 26 °C in the dark for 14 h.Fluorescence in transformed protoplasts was recorded and images were acquired with a confocal laser scanning microscope (TCS SP8, Leica).
2.5.Co-immunoprecipitation (Co-IP) assays
To generate the SPL-GFP and WOX-Flag plasmids, the fulllength coding regions of OsSPL10 and OsWOX3B were amplified by PCR and inserted separately into the vectors pBin-GFP and pYBA1142 (primers are listed in Table S1).The WOX-Flag and SPL-GFP, and WOX-Flag and pBin-GFP were co-introduced separately into two-week-old rice leaf protoplasts.After the protoplasts were cultured at 26 °C, 60 r min-1for 16 h, total protein was extracted with the Co-IP buffer (50 mmol L-1Tris-MES at pH 8.0,0.5 mol L-1sucrose,1 mmol L-1MgCl2,10 mmol L-1EDTA,5 mmol L-1DTT, and 1% (v/v) protease inhibitor).Immunoprecipitation was performed using GFP magnetic beads(LABLEAD),and the precipitated complexes were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and detected by immunoblotting with corresponding antibodies.
2.6.Dual-luciferase reporter (DR) assay
The 2.5-kb promoter region of HL6 and 2.1-kb promoter region of OsWOX3B were amplified and inserted into the pGreen II 0800-LUC vector to generate the reporter constructs ProHL6-LUC and ProOsWOX3B-LUC, respectively.The full-length coding regions of OsSPL10 and OsWOX3B were inserted separately into the pGreen II 62-SK vector (primers are listed in Table S1).The constructed recombinant plasmids were co-introduced into rice protoplasts as described [24].Firefly luciferase (luc) and Renilla luciferase(Rluc) activities were measured with the Dual-Luciferase Assay Kit (Vazyme) according to the manufacturer’s recommendations,and the relative LUC activity was calculated as the ratio luc/Rluc.At least three transient assay measurements were conducted for each assay.

Fig.1.Relative expression levels of OsSPL10, OsWOX3B, and HL6 in the shoots of seedlings of 3 mutants or 2 overexpression lines in comparison with the wild types.(A)CRISPR/Cas9 editing of OsSPL10 and OsWOX3B in ZH11 and R401,and T-DNA insertion of HL6 in ZH11.The gene framework of OsSPL10,OsWOX2B,and HL6 showing the coding region (filled boxes), UTRs (blank boxes), introns (horizontal lines), mutation sites (dotted line), T-DNA insertion site (blank triangle).(B–D) Relative expression levels of OsSPL10 (B), OsWOX3B (C) and HL6 (D) in Osspl10, hl6 and wild type ZH11, and in Oswox3b and wild type R401.Relative expression levels of OsWOX3B (E) and HL6 (F) in OsSPL10-overexpression lines OsSPL10-OE#24 and OsSPL10-OE#29 and wild type ZH11.Error bars represent SD (n = 3).**, P < 0.01 by Student’s t-test.
2.7.Electrophoretic mobility shift assay (EMSA)
To produce the recombinant proteins of GST-SPL10, GSTWOX3B, and GST-HD in E.coli, the coding regions of OsSPL10 and OsWOX3B and the homeobox domain (HD) of OsWOX3B were inserted separately into the pGEX-4 T-1 vector and expressed in the E.coli BL21 (DE3) strain.Recombinant proteins were purified according to the manufacturer’s instructions for Glutathione MagBeads (GenScript, Nanjing, Jiangsu, China).Oligonucleotide probes 1 and 3 containing GTAC motif and probe 2 containing TTAATAG motif for EMSA were synthesized by(Sunya, Fuzhou, Fujian, China).Labeled probes with Cy5(sulfo-cyanine 5) at the 3′end were used as binding probes and unlabeled probes were used as competitors.The sequences of primers and probes are presented in Table S1.The probes were incubated with the indicated amounts of recombinant proteins in a 20-μL reaction (100 mmol L-1Tris-HCl, pH 7.5,100 mmol L-1KCl, 50 mmol L-1MgCl2, 1 mmol L-1DTT,0.05 mg mL-1poly [dI-dC]) at 4 °C for 30 min.The DNA-protein complexes were separated on 6% (w/v) native polyacrylamide gels and images were captured with an Odyssey CLx Infrared Imaging System (LI-COR Biosciences).
2.8.Yeast one-hybrid (Y1H) assay
The full-length CDS region of OsSPL10 was introduced into the yeast expression vector pB42AD (Clontech), yielding the construct OsSPL10-pB42AD.The 2.1-kb promoter region and two truncated promoter regions of OsWOX3B were introduced into the yeast expression vector placzi2u(Clontech),yielding respectively the following constructs: proOsWOX3B-placzi2u, proOsWOX3B-1438-placzi2u, and proOsWOX3B-1360-placzi2u.The primers used are listed in Table S1.The fusion constructs were co-introduced into yeast strain EGY48.The transformed strains were grown on SD/-Trp/-Ura medium at 30 °C for 3 d and their transcriptional activities were measured on SD/-Trp /-Ura + X-β-Gal selective medium.
3.Results
3.1.OsSPL10 indirectly positively regulated OsWOX3B expression
To investigate the complete regulatory network and also reconfirm the reported regulatory relationships among OsSPL10,OsWOX3B and HL6, we used qRT-PCR to measure the relative expression levels of OsSPL10, OsWOX3B, and HL6 in the respective mutants Osspl10, Oswox3b, and hl6 in comparison with those in the corresponding wild types (ZH11 or R401).In Osspl10, the expression levels of the three genes were all reduced; in Oswox3b,the expression of OsWOX3B and HL6 was down-regulated,but that of OsSPL10 remained unchanged; in hl6, only HL6 was downregulated in expression, but OsSPL10 and OsWOX3B were unaffected (Fig.1B–D).The relative expression levels of OsWOX3B and HL6 were both increased in the OsSPL10-overexpression lines(Fig.1E, F).
These findings suggested that OsSPL10 positively regulates OsWOX3B expression and reconfirmed that OsSPL10 and OsWOX3B positively regulate HL6 expression[19,21].In other words,OsSPL10 acts upstream of OsWOX3B and HL6, and OsWOX3B acts upstream of HL6 in terms of transcription regulation.However, while OsSPL10 as a transcription factor can bind to the promoter of HL6 [21], it cannot bind to the promoter of OsWOX3B, as shown by our transient DR, EMSA and Y1H experiments (Fig.2).In the transient DR assay (Fig.2A), the luc/Rluc ratio using OsSPL10 as effector was not different from that of the negative control(Fig.2B), indicating that overexpression of OsSPL10 did not affect OsWOX3B transcription.In the EMSA assay, no signal of combination was detected between GST-OsSPL10 and probe 3, which carried the predicted OsSPL10-binding site (GTAC motif) in the OsWOX3B promoter region (Fig.2C, D).In the Y1H assay, in the presence or absence of OsSPL10, the two truncated promoter sequences of OsWOX3B were always inactive, whereas the fulllength OsWOX3B promoter was always active,suggesting that transcription was independent of OsSPL10 but activated by a yeast protein (Fig.2D, E).These results consistently indicated that OsSPL10 regulates OsWOX3B expression indirectly.
3.2.OsSPL10 physically interacted with OsWOX3B
OsWOX3B regulate the expression of HL6, and OsWOX3B and HL6 physically interact with each other through their homeobox and AP2 domains [19].Given that OsSPL10 regulated the expression of OsWOX3B (Fig.1B), we hypothesized that OsSPL10 physically interacts with OsWOX3B.
To test this hypothesis, we performed Y2H assays.Besides examining the full-length OsWOX3B and OsSPL10, we checked two truncated proteins (containing either HD or WUS domain) of OsWOX3B and three truncated proteins (OsSPL10-N-terminal,SBP domain and OsSPL10-C-terminal) of OsSPL10 (Fig.3A).The Y2H assays showed that both the full-length OsSPL10 and its Nterminal region interacted with the full-length OsWOX3B and its HD domain, but showed no interaction in other protein combinations (Fig.3B).This result suggested that OsSPL10 and OsWOX3B mutually interact through their N-terminal region and HD domain.
To determine whether the interaction between OsSPL10 and OsWOX3B also occurs in vivo, we performed a BiFC assay in rice protoplasts.The yellow fluorescent protein(YFP)fluorescence fully overlapped with the red fluorescence of OsH2B-mCherry in the nuclei of protoplast cells when OsSPL10-YFPn and OsWOX3BYFPc were co-expressed, but no YFP fluorescence signal was observed in negative controls (YFPn co-expressed with OsWOX3B-YFPc, or OsSPL10-YFPn co-expressed with YFPc;Fig.3C).These results confirmed that the OsSPL10–OsWOX3B interaction occurs in nuclei in vivo.
In Co-IP assays of OsSPL10 and OsWOX3B, recombinant OsSPL10-GFP was precipitated by OsWOX3B-Flag, but GFP alone was not (Fig.3D).Both the recombinant OsSPL10-GFP and GFP were precipitated by GFP, indicating that the GFP immune response system functioned normally.Thus, the Co-IP assays further confirmed the in vivo interaction between OsSPL10 and OsWOX3B.
These results indicated that OsSPL10 and OsWOX3B physically interact with each other.
3.3.OsSPL10 and OsWOX3B attenuate each other in promoting HL6 transcription
Based on the finding of physical interaction between OsSPL10 and OsWOX3B, we wondered how their interaction would affect the regulation of HL6 expression.To answer this question,we performed transient DR assays in rice protoplasts.In this system, the luc (firefly luciferase) reporter gene was driven by a 2.5 kb DNA fragment before the start codon ATG of HL6, which contained the core binding motif GTAC of OsSPL10 and the core binding motif TTAATGG of OsWOX3B (Fig.4B).Overexpression of OsSPL10 or OsWOX3B increased HL6 transcription compared with the negative control, but co-expression of OsSPL10 and OsWOX3B did not(Fig.4C).
The above results suggested that the OsSPL10-OsWOX3B interaction could hinder their binding to HL6 promoter.To test this hypothesis, we performed an EMSA experiment in vitro, using DNA fragments containing either the OsSPL10 (probe 1) or the HD (probe 2) binding site in the HL6 promoter region as probes.GST-OsSPL10 bound directly to the target probe 1,and the binding efficiency was significantly reduced in the presence of GSTOsWOX3B in the reaction,but not in the presence of GST tag alone(Fig.4D).Likewise, GST-HD bound directly to target probe 2, and the binding efficiency was significantly reduced in the presence of GST-OsSPL10 in the reaction, but not in the presence of GST tag alone (Fig.4E).
In summary, these results suggested that OsSPL10 and OsWOX3B attenuate each other’s ability to bind to the promoter and activate the transcription of HL6.
4.Discussion
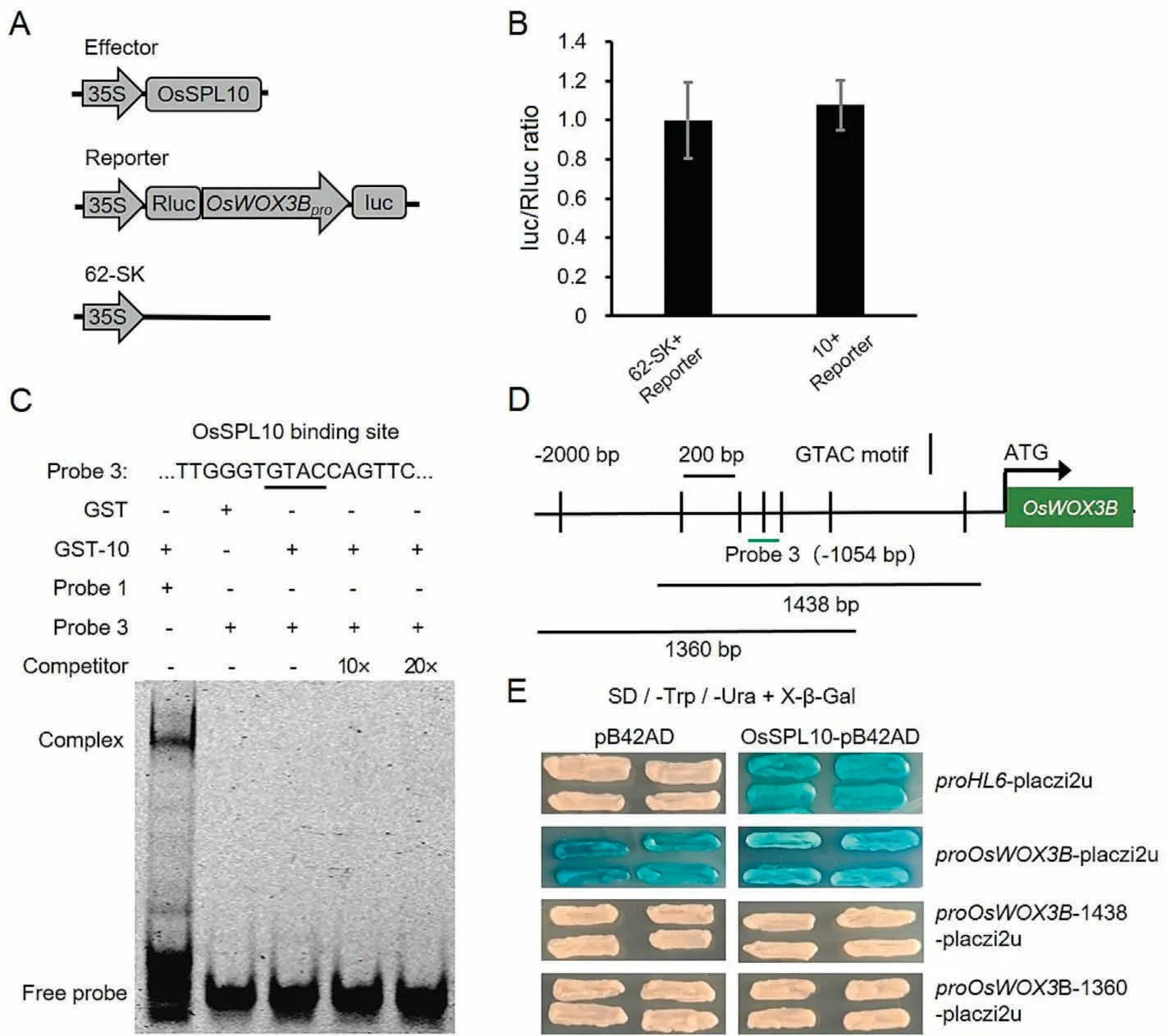
Fig.2.OsSPL10 could not bind to the promoter of OsWOX3B.(A) The schematic diagram of effector and reporter of double luciferase reporter experiment.Firefly luciferase gene driven by OsWOX3B promoter and Renilla luciferase gene driven by CaMV35S promoter were used as the reporter gene and the internal reference gene,respectively.(B)Ratio of luc to Rluc activity.The effector and the reporter were co-introduced into rice protoplasts and expressed transiently.62-SK and the reporter were co-introduced as negative control.10 represents 35S-OsSPL10.(C)EMSA of the DNA binding activities of OsSPL10 to the GTAC motif in the promoter of OsWOX3B.The 10×and 20×refer to the concentration of the competitive probe relative to that of the labeled probe.GST protein was used as negative control.The complex of GST-10 and probe 1 used as a positive control was the same as the complex used in Fig.4D.The symbols + and - indicate respectively the presence and absence of the corresponding proteins or probes.10 represents OsSPL10.(D)Schematic diagram of OsSPL10 binding motif GTAC and probe 3 position in the OsWOX3B promoter region,and two truncated promoters of OsWOX3B.(E)Y1H assay.Transformed cells were streaked on the selective medium SD/-Trp/-Ura+X-β-Gal.OsSPL10-pB42AD co-transformed with proHL6-placzi2u was used as positive control.pB42AD co-transformed with proHL6-placzi2u was used as negative control.
Trichome development can be roughly divided into two stages:initiation and elongation.OsWOX3B[10–12]and OsSPL10[16]positively control trichome development involving both initiation and elongation, while HL6 promotes only trichome elongation [19].These findings suggest that OsWOX3B and OsSPL10 function earlier than HL6 and must function upstream of HL6 in the regulatory pathway for trichome development.This hypothesis is consistent with the findings that both OsWOX3B [19] and OsSPL10 [21] positively regulate the expression of HL6.These findings were verified in the present study (Fig.1B, D, F).Given the finding that OsSPL10 also positively regulated the expression of OsWOX3B(Fig.1B,C,E),the order of the regulatory relationships among these three genes must be OsSPL10 →OsWOX3B →HL6.The finding that OsSPL10 did not bind to the promoter of OsWOX3B(Fig.2)suggests that OsSPL10 regulates only indirectly the expression of OsWOX3B.
A transcriptional regulation mechanism similar to that involving the SPL and WOX genes for trichome development has been found in maize.There are two protein orthologs of OsWOX3B in maize: ZmWOX3A and ZmWOX3B.Zmspl10/14/26 triple mutants exhibit complete hairlessness,and these ZmSPLs promote trichome formation by positively regulating the expression of ZmWOX3A[25].WOX transcription factors have conserved functions among plant species [26], and OsSPL10 is the ortholog of ZmSPL14 in rice[25].It thus appears that the relationship of SPL gene(s)regulating the expression of WOX gene(s)for trichome formation represents a mechanism conserved across grass species.However, the finding[25] that ZmSPL10/14/26 binds to the promoter of ZmWOX3A to upregulate its expression is in contrast to that of the interaction between OsSPL10 and OsWOX3B in rice found in the present study.
Besides the relationships of transcription regulation, OsSPL10,OsWOX3B and HL6 physically interact at the protein level.OsWOX3B and HL6 can combine into a heterologous complex that increases the binding of HL6 to the promoter of the target gene OsYUCCA5 to increase auxin synthesis for trichome elongation[19].OsSPL10 did not interact with OsWOX3B and HL6 in yeast cells [21], nor did the present study reveal physical interaction between OsSPL10 and HL6.However, OsSPL10 physically interacted with OsWOX3B in the present study (Fig.3).Thus, both HL6 and OsSPL10 physically interact with OsWOX3B.The finding that OsWOX3B interacted with the two proteins both through its homeobox domain, which combined with the AP2 domain of HL6[19] and the N-terminal region of OsSPL10 (Fig.3B), indicates the importance of the homeobox domain in OsWOX3B.
Unlike the interaction between OsWOX3B and HL6,which facilitates the function of HL6 as a transcription factor, the OsSPL10-OsWOX3B interaction does not improve the function of either protein.In the transient analyses, overexpression of either OsSPL10 or OsWOX3B increased HL6 transcription compared with the negative control, but co-overexpression of the two genes did not (Fig.3B).Instead,the EMSA assay revealed that OsSPL10 and OsWOX3B even reduced each other’s ability of binding to the promoter of HL6(Fig.3C, D).These results suggested that the physical interaction between OsSPL10 and OsWOX3B may hinder both proteins from activating the expression of HL6.This might provide a way to regulate the expression level of HL6.It is known [19] that HL6 regulates trichome elongation in a dosage-dependent manner, with higher HL6 expression leading to longer trichomes.Given that both OsSPL10 and OsWOX3B alone promote HL6 expression and OsSPL10 upregulates OsWOX3B expression, it is likely that the expression level of HL6 would be very high and therefore trichomes would be very long if there were not a balancing regulation mechanism.The OsSPL10–OsWOX3B interaction could be such a mechanism,enabling trichomes to develop within a suitable length range.

Fig.3.Detection of physical interaction between OsSPL10 and OsWOX3B.(A) Schematic diagram of the protein structures of OsWOX3B and OsSPL10.The conserved homeobox,WUS,and SBP domains are indicated.(B)Y2H assay.Transformed cells were spotted on the control medium DDO(SD/-Leu/-Trp)and selective medium QDO(SD/-Leu/-Trp/-His/-Ade).(C) BiFC assays in rice protoplasts.YN, N-terminal region for YFP; YC, C-terminal region for YFP; SPL10 and WOX3B represent OsSPL10 and OsWOX3B,respectively.mCherry, the fusion protein OsH2B-mCherry, which is a marker with red fluorescence localized in the nucleus.Scale bars, 7 μm.(D) Co-IP assays in rice protoplasts.Precipitates were detected by Western blotting with anti-GFP (recognizing OsSPL10-GFP) and anti-FLAG (recognizing OsWOX3B-FLAG)antibodies respectively.Protein size(kD)is shown at the left of the gel.SPL10 and WOX3B represent OsSPL10 and OsWOX3B,respectively.IP and IB represent respectively immunoprecipitation and tag antibodies in Western blot.
Incorporating the findings of this study with those on the relationships among OsSPL10, OsWOX3B and HL6 reported previously,we propose a model (Fig.5) of rice trichome development regulated by these three genes.OsSPL10 positively regulates the expression of OsWOX3B to initiate trichomes with the mediation of other transcription factors.Both OsSPL10 and OsWOX3B activate the transcription of HL6 by binding to its promoter, increasing trichome elongation.The activating effects of OsSPL10 and OsWOX3B on HL6 transcription are not additive but mutually attenuating owing to the formation of a OsSPL10-OsWOX3B dimer,which cannot activate HL6 expression.The balance of abundance of the heterodimer and the two monomers in the cell provides a mechanism for adjusting the expression of HL6 to regulate trichome growth to a suitable length.
The newly reported LHL1 gene governing rice trichome development does not encode a transcription factor but a plant-specific,highly conserved nuclear protein that contains an RNaseH-like domain.Loss of LHL1 function reduced the expression of OsWOX3B and HL6 [20].This finding suggested that LHL1 affects trichome development by regulating the expression of OsWOX3B and HL6.Whether LHL1 also regulates OsSPL10 expression was not mentioned by these authors.We found that LHL1 expression was not altered in either the Osspl10 mutant or the OsSPL10-overexpression line compared with the wild type ZH11 (Fig.S1),suggesting that the expression of LHL1 is not regulated by OsSPL10.Further study might reveal whether the expression of OsSPL10 is affected by LHL1.If so, then LHL1 is possibly located in the same pathway and upstream of the three transcription-factor genes.If not, then it is possible that OsSPL10 and LHL1 represent different pathways, regulating OsWOX3B and HL6 independently.

Fig.4.OsSPL10 and OsWOX3B attenuate each other in binding to the promoter to activate the transcription of HL6.(A) Schematic diagram of OsSPL10 binding motif GTAC and OsWOX3B binding motif TTAATAG in the HL6 promoter region (-2.5 kb) and the probes used for EMSA.(B) The schematic diagram of effector and reporter of double luciferase reporter experiment.Firefly luciferase gene driven by HL6 promoter and the Renilla luciferase gene driven by CaMV35S promoter were used as the reporter gene and the internal reference gene,respectively.(C)Ratio of luc to Rluc activity.The effector and the reporter were co-introduced into rice protoplasts and expressed transiently.62-SK and the reporter were co-introduced as negative control.10 and 3B represent 35S-OsSPL10 and 35S-OsWOX3B, respectively.Different lowercase letters indicate differences at P<0.05 detected by one-way ANOVA.(D)and(E)EMSA of the DNA binding activities of OsSPL10 to the GTAC motif(D)and OsWOX3B to the TTAATAG motif(E)in the promoter of HL6.The 10×and 20×refer to the concentration of the competitive probe relative to that of the labeled probe.GST protein was used as negative control.The symbols + and - indicate respectively the presence and absence of the corresponding proteins or probes.10 and 3B represent OsSPL10 and OsWOX3B, respectively.

Fig.5.A schematic model of rice trichome development regulated by OsSPL10,OsWOX3B and HL6.See text for the description of the model.Meanings of symbols:○and?,proteins;→,production;,activation of transcription;,indirect activation of transcription;, leading to.
5.Conclusions
OsSPL10 positively regulated the expression of OsWOX3B and HL6 in trichome development in rice.OsSPL10 physically interacted with OsWOX3B.This interaction attenuated the abilities of both proteins to bind to the promoter of HL6 to activate its transcription.
CRediT authorship contribution statement
Qiaoling Liao:Investigation, Writing - original draft.Xinle Cheng:Investigation, Writing - original draft.Tong Lan:Writing- editing.Xiaokuan Guo:Investigation.Zilong Su:Investigation.
Xiaoxiao An:Investigation.Yali Zheng:Investigation.Haitao Cui:Supervision.Weiren Wu:Conceptualization,Writing-review& editing.Tao Lan:Funding acquisition, Supervision, Conceptualization, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31671668), Natural Science Foundation of Fujian Province (2021J01076), International Atomic Energy Agency Coordinated Research Project (D23031–22287), and Key Program of Science and Technology in Fujian Province (2020NZ08016).We thank Professor Sibin Yu of Huazhong Agricultural University for mutant hl6 seeds.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.05.012.
- The Crop Journal的其它文章
- Ectopic expression of OsNF-YA8, an endosperm-specific nuclear factor Y transcription-factor gene, causes vegetative and reproductive development defects in rice
- Mechanisms of autophagy function and regulation in plant growth,development, and response to abiotic stress
- ZmDRR206 functions in maintaining cell wall integrity during maize seedling growth and defense response to external stresses
- The plasmodesmata-associated β-1,3-glucanase gene GhPdBG regulates fiber development in cotton
- The MabHLH11 transcription factor interacting with MaMYB4 acts additively in increasing plant scopolin biosynthesis
- Evolutionary genetics of wheat mitochondrial genomes

