Effects of Main Nitrogen-Regulating Gene AreA on Growth, Pathogenicity, and Fumonisin Synthesis of Fusarium proliferatum
Effects of Main Nitrogen-Regulating Geneon Growth, Pathogenicity, and Fumonisin Synthesis of
Supplemental data
File S1. Methods
Fungal strains and culture conditions
The FP9 strain ofwas isolated from samples infected by rice spikelet rot disease (RSRD) and used as the wildtype (WT) strain. This strain produces large amounts of fumonisin and is highly pathogenic(Sun et al, 2019).Its colony morphology was compared onpotato dextrose agar (PDA) media, and the radial growth was determined by measuring colony diameters after 5 d growth on PDA and minimal medium (containing sucrose 30 g, KH2PO41 g, MgSO4?7H2O 0.5 g, KCl 0.5 g, and a 0.2 mL solution of trace elements). The various N sources tested included 12 mmol/L of sodium nitrate (NaNO3), 120 mmol/L of NaNO3, 6 mmol/L of ammonium nitrate (NH4NO3), 60 mmol/L of NH4NO3, 6 mmol/L of glutamine (Gln), and 60 mmol/L of Gln, each in a constant volume of 1 L. The trace elements solution included citric acid 5 g, zinc sulfate heptahydrate (ZnSO4?7H2O) 5 g, iron sulfate heptahydrate (FeSO4?7H2O) 4.75 g, copper sulfate pentahydrate (CuSO4?5H2O) 250 mg, magnesium sulfate monohydrate (MnSO4?H2O) 50 mg, boric acid (H3BO3) 50 mg, and sodium molybdate dihydrate (Na2MoO4?2H2O) 50 mg in a constant volume of 100 mL was used to determine the effect of different N sources on growth. They were added to rice grain (RG) media (20 g rice grains and 20 mL water per bottle) to measure the effect of different N sources (12 mmol/L of NaNO3, 120 mmol/L of NaNO3, 12 mmol/L of NH4Cl, 120 mmol/L of NH4Cl, 6 mmol/L of NH4NO3, 60 mmol/L of NH4NO3, 6 mmol/L of Gln, and 60 mmol/L of Gln) on fumonisin biosynthesis. Fungal genomic DNA was isolated from mycelia grown in liquid YEPD media (containing 3 g yeast extract, 10 g peptone, and 20 g dextrose per 1 L), and conidia were produced for inoculum by growing the fungi in liquid mung bean (MB) media [10 g mung bean () with H2O added to bring the volume to 1 L].
Deletion and complementation of AreA
In this study, the evolutionary trend ofhomologs in the phylogenetic tree was generally consistent with the evolutionary trend of the species, suggesting that thegenes in the phylogenetic tree were the direct homologs of AreA/Nit2 in other ascomycetes. Thus, their functions should be consistent with or similar to those of the AreA/Nit2 family members.The deletion constructs forwere engineered utilizing approximately 1.5–2.0 kb upstream and downstream regions that flank the coding region. The upstream/downstream regions and hygromycin B resistance gene () were amplified by PCR from genomic DNA and plasmid DNA (pCPXHY2GFP), respectively. The three fragments were then integrated into the plasmid pBluescript II SK using a multi-fragment recombination kit (Yeasen Biotechnology Co., Ltd., Shanghai, China). Complementation constructs forfirst amplified a fragment that contained the promoter region that was 1.5 kb upstream of thefragment, the full length of thefragment, and the terminator region 300 bp downstream. The resistance marker G418 was then amplified, and the two DNA fragments were finally fused for complementation experiments. Table S1 lists the primers used to amplify each region of each gene. The resulting vectors were transformed separately into the WT and mutant strainΔas described by Sun et al(2019). PCR was used to determine three independently isolated deletion mutants of thegene analyzed in this study, which were then selected for Southern blot analysis to confirm the deletion of targetedgene.Southern blotting were performed by isolating the genomic DNA, digesting it withRV, separating the digested DNA using 0.8% agarose gel electrophoresis, and blotting it onto a nylon membrane. In the engineered strains, HygR and G418 were labeled with digoxigenin (DIG) using the DIG-High Prime Labeling and Detection Kit II (Roche, Penzberg, Germany), and hybridized to the Southern blotting(Glenn et al, 2008). The fragments were detected and visualized according to the manufacturer’s instructions.
Analysis of fumonisin B1 (FB1) production
The concentration of the conidial solution of each strain was adjusted to approximately 3 × 106spores/mL, and 1 mL of this suspension was inoculated into different N sources that were utilized in theRG media (20 g rice grains and 20 mL water per bottle). The strains were cultured on sterilizedRG at 28 °C for 7 dto assess the production of FB1. High performance liquid chromatography tandem mass spectrometry (HPLC-MS/MS) was used to measure the FB1contentin the culture extracts as described by Li et al(2014). The sample was extracted with a methanol-water-acetic acid (74:25:1) solution and cleaned using a strong anion exchange column (SAX). FB1was completely separated on a ZORBAX Extend-C18column (150 mm × 2.1 mm, 1.8 μm) with a gradient elution using 0.1% acetic acid-water and acetonitrile as the mobile phases, respectively. The eluents were detected by positive electrospray ionization mass spectrometry under select reaction monitoring mode. Ergosterol is an important component of fungal cell membranes and is often used as a standard for determining the biomass of living fungi.Ergosterol analysis was performed on a high-performance liquid chromatography system with UV detection at 282 nm and a 4.6-U octadecylsilyl (ODS) column. Compounds were eluted with 100% methanol at a flow rate of 1.0 mL/min as described by Bluhm and Woloshuk (2005). The experiment was independently repeated with three biological replicates.
Virulence assay
The susceptible rice (subsp.) cultivar Xiushui 134 was used for the pathogenicity assays. We modified the method described by Sun et al (2018) to develop ourassay. Briefly, three panicles of rice at the pollen cell meiosis to the maturity stages were inoculated with the WT and mutant strains at a concentration of 5×105spores/mL. Infected rice plants were placed in a growth chamber with a controlled temperature (28 oC), relative humidity (80%), and light cycling (16 h light and 8 h dark). Rice plants inoculated with sterile water served as the control. The panicle was sampled and observed for symptoms of (RSRD) 14 d after inoculation. The RSRD disease index was used to determine the index for panicle infection as defined by Huanget al (2011b). The grades were defined as follows: grade 0, no grains infected; grade 1, 0.1%–10.0% grains infected; grade 3, 10.1%–25.0% grains infected; grade 5, 25.1%–50.0% grains infected; grade 7, 50.1%–75.0% grains infected; and grade 9, greater than 75.1% grains infected.The disease index was calculated according to the following formula:
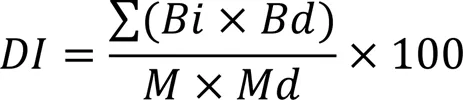
whereis the disease index,is number of panicles with grade,is the individual grade value (0 to 9),is the total number of observed panicles, andis the maximum grade value.
qRT-PCR expression analysis
For each strain, the conidial solution described for FB1production was inoculated into 50 mL of potato dextrose media and incubated in a rotary shaker (180 r/min) at 28 oC for 3 d. The mycelia were collected, and 1 g (wet weight) was re-inoculated into 50 mL of GAPL media (3 g KH2PO4, 0.3 g MgSO4, 5 g NaCl, 6 mmol/L glutamine, and 20 g sucrose in 1 L). We then collected the mycelia at 48 h for qRT-PCR analysis to assess the expression ofgenes. The primers used for qRT-PCR were described by Sun et al (2019).
Total RNA samples were isolated from the mycelia of strains grown in GAPL media for 48 h using the RNAiso Plus reagent (Takara Bio, Inc., Shiga, Japan). First-strand cDNA was synthesized using a PrimeSciptTMRT Reagent Kit with gDNA Eraser (Takara Bio, Inc. Tokyo, Japan), following the manufacturer’s instructions. The β-tubulin gene (was used as an endogenous control to normalize differences in the quantity of mRNA owing to differing amounts of total RNA. The levels of expression of each gene were calculated using the 2-ΔΔCtmethod. Data from three biological replicates were used to calculate the mean and standard deviation. The statistical analysis was processed and plotted in Microsoft Excel 2003 (Redmond, WA, USA) and GraphPad 6 (San Diego, CA, USA). The Duncan method in SAS 9.2 statistical software (Cary, NC, USA) was used to test for significant differences.
Bluhm B H, Woloshuk C P. 2005. Amylopectin induces fumonisin B1production byduring colonization of maize kernels., 18(12): 1333–1339.
Glenn A E, Zitomer N C, Zimeri A M, Williams L D, Riley R T, Proctor R H. 2008. Transformation-mediated complementation of agene cluster deletion inrestores both fumonisin production and pathogenicity on maize seedlings., 21(1): 87–97.
Huang S W, Wang L, Liu L M, Tang S Q, Zhu D F, Savary S. 2011. Rice spikelet rot disease in China: 2. Pathogenicity tests, assessment of the importance of the disease, and preliminary evaluation of control options., 30(1): 10–17.
Li Z X, Chen X L, Cao Z Y, Cao X L, Gong J D, Zhu Z W. 2014. Determination of fumonisins in cereals using liquid chromatography-tandem mass spectrometry., 33(2): 167–172. (in Chinese with English abstract)
Sun L, Wang L, Liu L M, Hou Y X, Li Q Q, Huang S W. 2018. Screening for strains of rice spikelet rot disease pathogenic fungus with high fumonisin production and strong pathogenicity., 32(6): 610–616. (in Chinese with English abstract)
Sun L, Wang L, Liu L M, Hou Y X, Xu Y H, Liang M Q, Gao J A, Li Q Q, Huang S W. 2019. Infection and colonization of pathogenic fungusin rice spikelet rot disease., 26(1): 60–68.
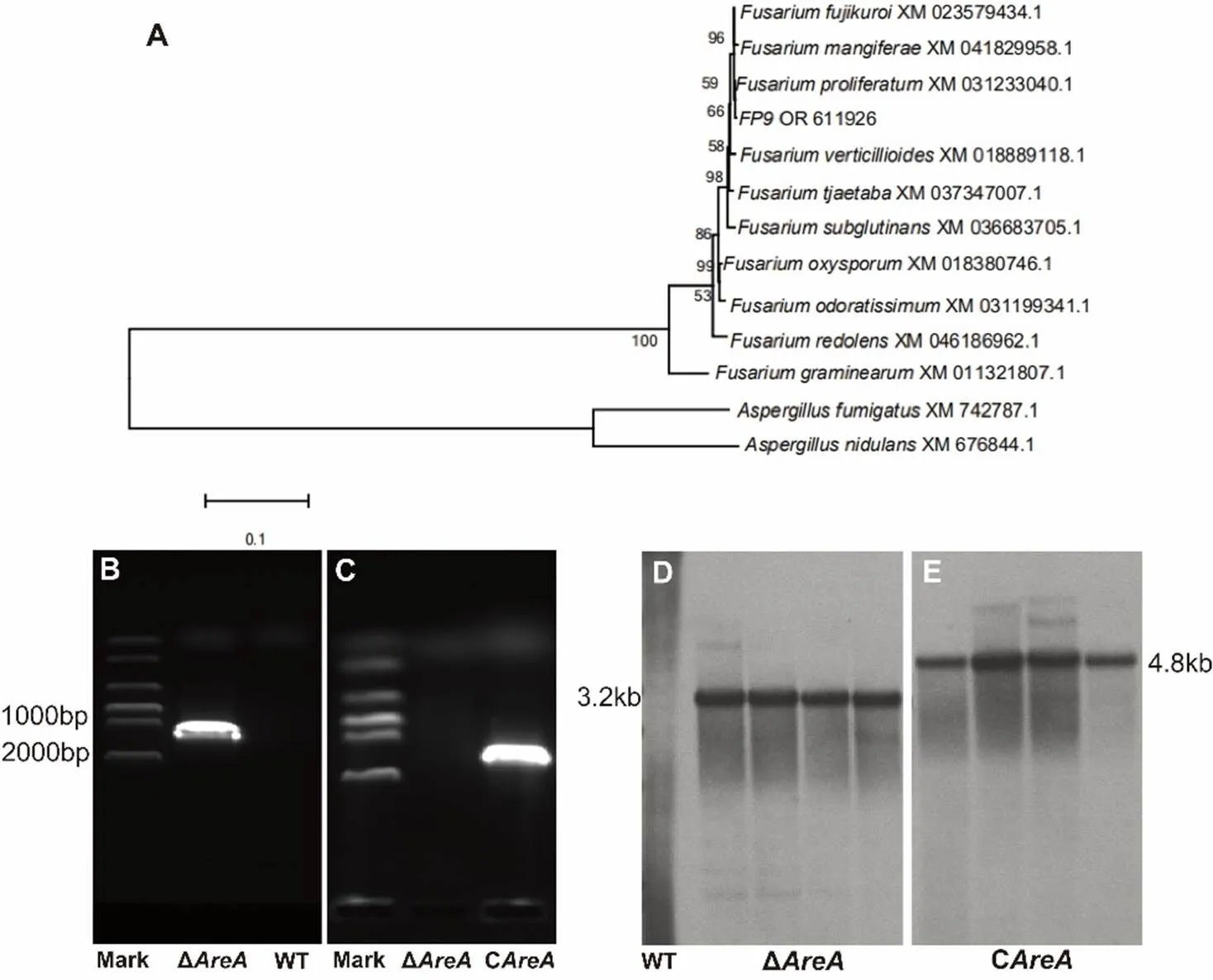
Fig. S1. Phylogenetic tree of(A), reverse transplant PCR (B and C) and Southern blot (D and E) detection of Δand C.
WT, Wild type; Δ, Deletion mutant; C, Complementation mutant.
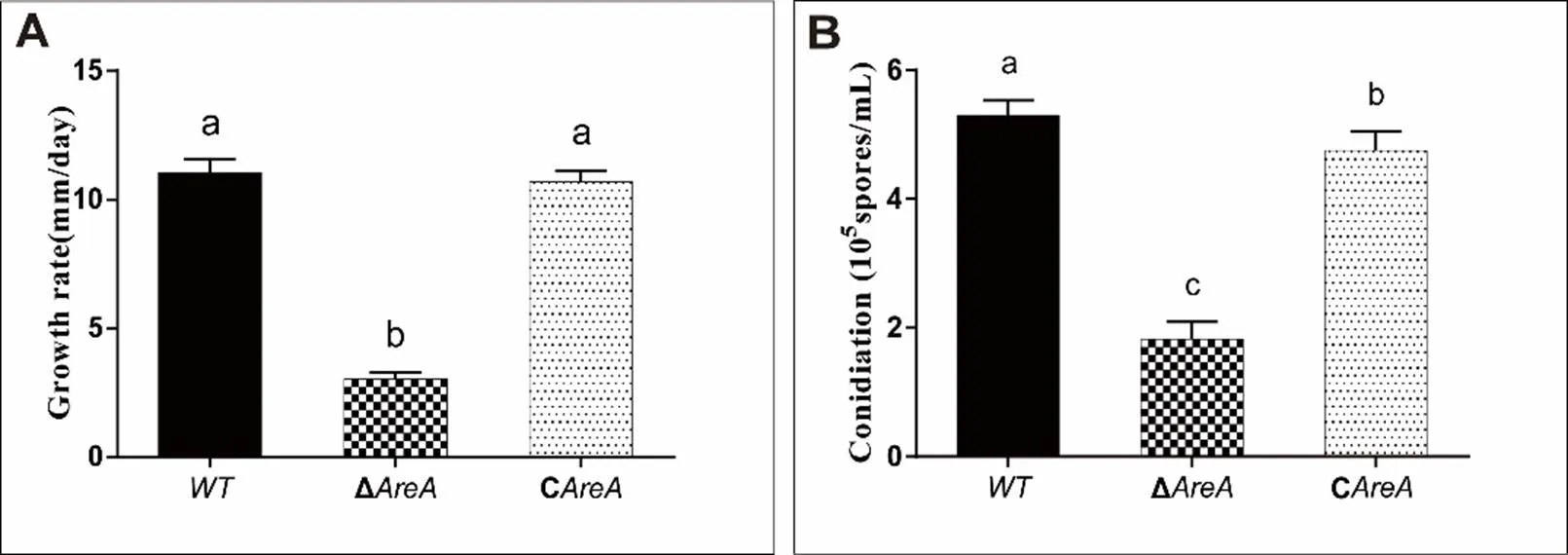
Fig. S2. Growth rates of wild type (WT), Δ, and Con potato dextrose agar media for 5 d (A) and conidiation in liquid mung bean media for 3 d (B).
WT, Wild type; Δ, Deletion mutant; C, Complementation mutant.
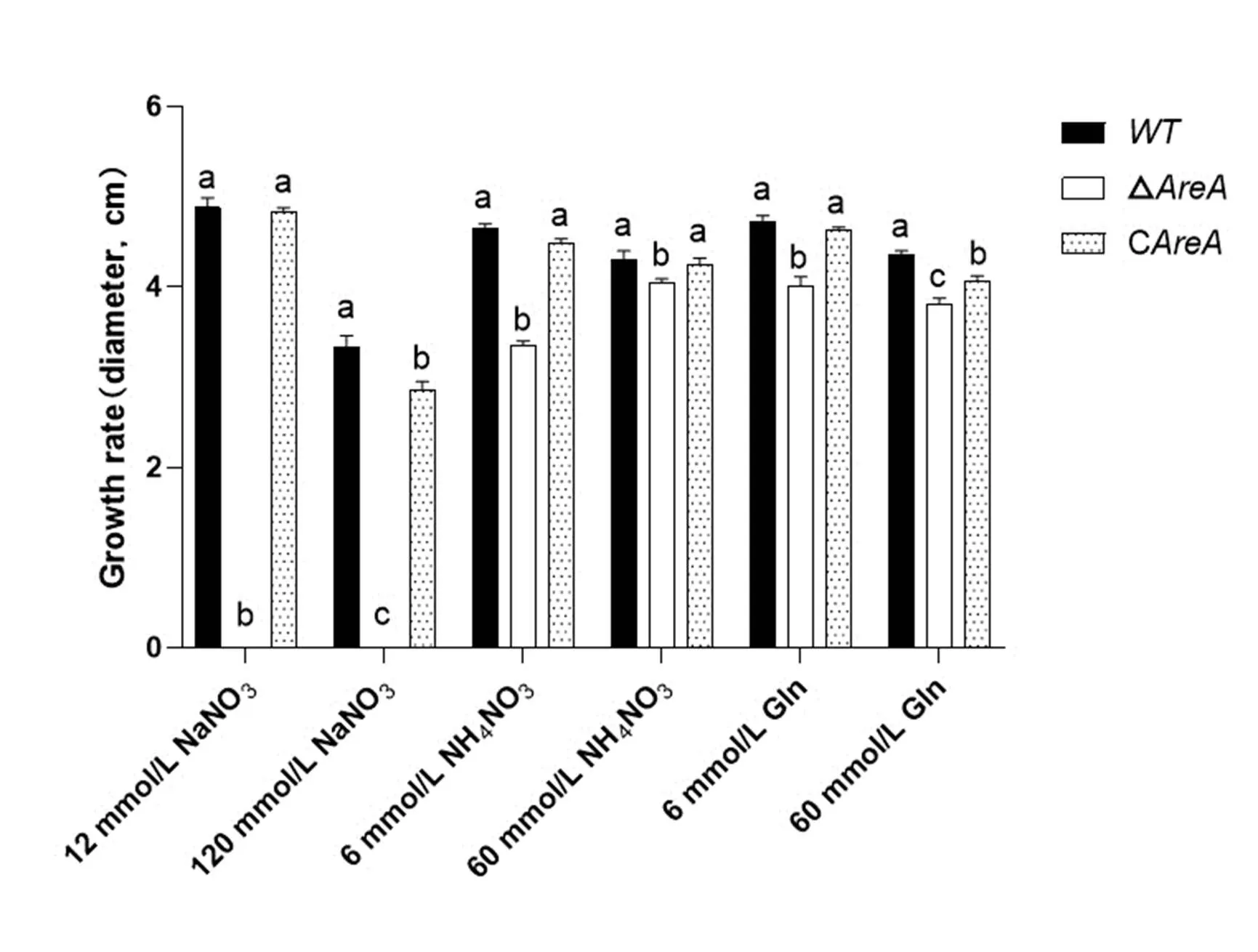
Fig. S3. Growth rates of wild type (WT), Δand Cstrains on minimal media with different nitrogen sources for 5 d.
WT, Wild type; Δ, Deletion mutant; C, Complementation mutant.
Table S1.Primers used in this study.

Primer nameSequenceProduct (bp) AreA-FD-FACTCGGTTACATCCATCG2100 AreA-FD-RCATTCATTGTTGACCTCCACTAAAGCCAGTAGGGAAACTTATC AreA-BK-FGGGCAAAGGAATAGAGTAGATGAGCCCTGAACAGAAATCCA1978 AreA-BK-RCACTTAGTACCCACGAGCC CAreA-FAGCAGGCAAAAGCAATCCCT5106 CAreA-RAGCATTGATGTGTTGACCTCCATTCCTCGACAACATGCAGGTTTT Neo-FAAAACCTGCATGTTGTCGAGGAATGGAGGTCAACACATCAATGCT1363 Neo-RTCAGAAGAACTCGTCAAGAAG AreA-YZ-FTATTCACAACGCTCCCACTC1092 AreA-YZ-FCCCAAAGCATCAGCTCATC CAreA-YZ-FCAGGAGAGGCAAAGTTGATAGG1445 CAreA-YZ-FGGGTTGAGTTGGTGACGGAT HygR-FTAGTGGAGGTCAACAATGAAT1357 HygR-RCATCTACTCTATTCCTTTGCC RTFUM21-FCGACTGCCAGTATAAAGCC122 RTFUM21-RGTAGCGTAACAGTTTGAGGAG RTFUM1-FCCAACTCTTCTTCCCTGCTA119 RTFUM1-RCACCCTCTACCTCCCACA RTFUM6-FCTGGAAAGTATGCGGTCAA101 RTFUM6-RGCAGAACTCATCAGCGTCA RTFUM8-FGCGGAACGAGAAATAGTGA117 RTFUM8-RTGCTGGGTTGAAAGGGAG RTTUB-F2ACATCCAGACAGCCCTTTGTG106 RTTUB-R2AGTTTCCGATGAAGGTCGAAGA
- Rice Science的其它文章
- Rice Variety Classification Based on Optimized Near-Infrared Spectral Classification Model
- Effects of Nitrogen-Regulating Gene AreA on Growth,Pathogenicity,and Fumonisin Synthesis of Fusarium proliferatum
- Rice Husk at a Glance:From Agro-Industrial to Modern Applications
- Smart Farming for Sustainable Rice Production:An Insight into Application,Challenge, and Future Prospect
- A β-Carotene Ketolase Gene NfcrtO from Subaerial Cyanobacteria Confers Drought Tolerance in Rice
- OsbZIP01 Affects Plant Growth and Development by Regulating OsSD1 in Rice