Haplotype variation and KASP markers for SiPSY1 – A key gene controlling yellow kernel pigmentation in foxtail millet
Rongjun Zuo,Ynyn Zhng,Yning Yng,Chunfng Wng,Hui Zhi,Linlin Zhng,Sh Tng,Ynn Gun, Shunguo Li, Ruhong Cheng, Zhonglin Shng, Gunqing Ji,*, Xinmin Dio,,,*
a Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
b Crop Research Institute, Shandong Academy of Agricultural Sciences, Jinan 250100, Shandong, China
c Institute of Millet Crops, Hebei Academy of Agricultural and Forestry Sciences, Shijiazhuang 050035, Hebei, China
d College of Life Sciences, Hebei Normal University, Shijiazhuang 050024, Hebei, China
Keywords:Carotenoids Lutein Zeaxanthin Marker assisted selection Setaria italica
ABSTRACT Carotenoid biosynthesis and accumulation are important in determining nutritional and commercial value of crop products.Yellow pigmentation of mature kernels caused by carotenoids is considered a vital quality trait in foxtail millet, an ancient and widely cultivated cereal crop across the world.Genomic regions associated with yellow pigment content(YPC),lutein and zeaxanthin in foxtail millet grains were identified by genome-wide association analysis (GWAS), and SiPSY1 (Phytoene synthase 1 which regulates formation of the 40-carbon backbone of carotenoids) was confirmed as the main contributor to all three components by knockout and overexpression analysis.SiPSY1 was expressed in seedlings,leaves,panicles,and mature seeds,and was subcellularly localized to chloroplasts.Transcription of SiPSY1 in 15 DAP immature grains was responsible for YPC in mature seeds.Selection of SiPSY1 combined with increased YPC in mature grains during domestication of foxtail millet was confirmed.Haplotype analysis suggested that expression level of SiPSY1 could be a selection target for future breeding programs, and a KASP marker was developed for selection of favorable SiPSY1 alleles in breeding.The results of this work will benefit nutritional and commercial improvement of foxtail millet varieties, as well as other cereal crops.
1.Introduction
Carotenoids are a class of C40 isoprenoid pigments with more than 600 structures synthesized by plastids of photosynthetic organisms,including plants,bacteria and fungi[1,2].In plants,carotenoids are essential components regulating plant growth and partially form the antioxidant system in seeds [3].For humans,dietary carotenoids are important precursors of vitamin A and retinoid compounds essential for growth[4,5].The carotenoid biosynthetic pathway has been considered a target of breeding and metabolic engineering efforts in crop and fruit species, including rice (Oryza sativa L.) [6], maize (Zea mays L.) [7], wheat (Triticum aestivum L.) [8], pepper (Capsicum frutescens L.) [9] and cucumber(Cucumis sativus L.) [10].
Biosynthesis of carotenoids begins with formation of a 40-carbon backbone mediated by phytoene synthase,and four further biosynthetic steps lead to lycopene [11].Lycopene could be recycled by lycopene β-cyclase (LCYB) and lycopene ε-cyclase (LCYE)to form α-carotene.Zeinoxanthin is produced by hydroxylation of a β-ring whereas lutein is produced by hydroxylation of the εring of α-carotene.Moreover, lycopene can also be recycled by LCYB to form β-carotene, which is hydroxylated for biosynthesis of β-cryptoxanthin and zeaxanthin [12].Chromosomal ploidy[13]and dosage of phytoene synthase 1(PSY1 or Y1)in maize kernel endosperm was positively correlated with carotenoid content[14,15].In contrast, rice endosperm does not accumulate carotenoids, although three homologous PSY genes have been identified in the rice genome [2].In Arabidopsis thaliana, only one PSY was detected and was considered an important regulator in carotenoid biosynthesis [16].Moreover, genomic variation in PSY1 in grass species, including maize, teosinte (Zeamexicana L.), tripsacum(Tripsacum spp.), coix (Coix spp.), and sorghum (Sorghum bicolor L.), and a high level of genomic diversity within the PSY1 coding region was identified in both traditional landraces and improved cultivars [17].This suggests that genomic variation in PSY1 in cereal gene pools could be selected for breeding of higher quality cereal crop varieties.In addition, genome-wide association studies of kernel color traits in maize revealed that natural variation in PSY1,zep1,dxs2 and lcyE are responsible for carotenoid biosynthesis and retention in maize grain[18].This enables further enhancement of yellow pigment content in cereal grains through traditional breeding approaches.
Foxtail millet[Setaria italica(L.)Beauv.],an ancient crop species that originated from China nearly 11,500 years ago [19] is still widely cultivated for grain and forage in warm and temperate regions of the world to date [20].Yellow pigment is an important component determining appearance and nutritional quality of foxtail millet grain,and carotenoids are mainly responsible[21].Large scale identification of carotenoid content in kernels of foxtail millet accessions revealed wide variation from 1.91 to 28.54 mg kg-1[22], indicating potential feasibility of improving yellow pigment levels in foxtail millet grains.Zeaxanthin and lutein are the main carotenoids in foxtail millet.An evaluation of two hundred foxtail millet accessions Liu et al.[23] showed that zeaxanthin content was higher than lutein, with zeaxanthin concentration averaging 8.6 mg kg-1and lutein concentration averaging 3.1 mg kg-1.Moreover,kernel yellow pigment content in Chinese foxtail millet accessions varied with geographical distribution and length of the growth period[22].Wang et al.[24]mapped the white kernel gene on chromosome 4 and the grey kernel gene on chromosome 6 using a set of primary foxtail millet trisomic lines.Li et al.[25]established a link between SiPSY1 and variation in grain color by‘b*’ analysis.Nevertheless, genetic mapping of genes controlling the accumulation of yellow grain pigmentation in foxtail millet remains unclear.
The genome sequence of Setaria has been assembled [26,27].Natural populations for genome wide association study (GWAS)in Setaria[25,28,29]were established for identification of genomic loci contributing to economically important traits.This research has made foxtail millet an emerging model for genetic deciphering important morphological and agronomic traits in cereal species,especially in regard to C4photosynthesis and tolerance to abiotic stress [30,31].To date, the genetic control of carotenoids biosynthesis in foxtail millet grains remains unclear which has hindered breeding of this ancient crop species.
In this study, genetic regulators of carotenoid accumulation in foxtail millet were identified through GWAS, and further characterized by studies of allelic variation.Superior alleles identified in this trial will be useful in developing markers for marker assisted selection (MAS) in this crop species.Results of this work will promote genetic dissection of nutritional characters in foxtail millet to enhance quality traits of this ancient crop species.
2.Materials and methods
2.1.Genome-wide association analysis of yellow pigment content in foxtail millet grain
A previously published panel of foxtail millet accessions [28]was used in GWAS.A total of 390 foxtail millet landrace accessions derived from different eco-regions of foxtail millet field productions were sampled from previous studies [28] as mini collections that covered all typical growth areas of the whole panel and were cultivated during 2013 growth period at Taiyuan, Shanxi, China(37.5°N,112.6°E,altitude 780 m)and 2014 growth season at Jinan,Shandong, China (36.4°N, 117°E, altitude 24 m), respectively(Fig.S1).In each trial, each accession was grown in a 3 m, 2-row plot with a 40 cm row spacing.Five individuals were randomly selected and the harvested grain was bulked for YPC measurement measured using high performance liquid chromatography (HPLC)following Yang et al.[22]with three technical replicates.BLUE values for grain YPC,lutein and zeaxanthin contents in each accession were calculated using the R package and GWAS analysis was conducted using both BLUE and individual site values.Genotypic data previously reported by Jia et al.[28] was used for GWAS analysis.In this trial,726,080 SNPs distributed across all nine chromosomes(72,497 on chromosome 1; 97,363 on chromosome 2; 93,457 on chromosome 3;42,765 on chromosome 4;73,329 on chromosome 5;75,717 on chromosome 6;81,198 on chromosome 7;93,339 on chromosome 8, and 96, on chromosome 9) were used for further analysis.Association mapping was conducted according to the pipeline in the TASSEL 5 package [32] with WeightedMLM algorithms to detect loci significantly associated with yellow pigment content, using a mixed linear model (PCA + K).The wholegenome significance cutoff was set to be 10-7based on permutation tests described previously [33].Candidate genes contributing to yellow pigment content were identified in genomic linkage disequilibrium (LD) blocks (average ± 50 kb in for the foxtail millet genome) surrounding the most significant SNP, based on genomic annotations of reference Yugu 1 genome (https://phytozome.jgi.d oe.gov/pz/portal.html#!info?alias=Org_Sitalica) [26].
2.2.Functional verification of SiPSY1 in foxtail millet
To generate a SiPSY1 knockout construct, oligonucleotides applied for targeted mutagenesis were designed with online software (https://cbi.hzau.edu.cn/crispr/) and cloned into CRISPR/Cas9 vector pYLCRISPR/Cas9-MH.To generate a SiPSY1 overexpression vector, the target sequence of SiPSY1 was obtained by PCR amplification of genomic DNA from Yugu 1, and then ligated into the vector pCAMBIA-1305 (Cambia) using an In-Fusion Advantage PCR Cloning Kit (Clontech).Finally, all plasmids were separately transformed into calli induced from mature seeds of foxtail millet cultivar ‘Ci846’ using the Agrobacterium tumefaciens–mediated(EHA105) transformation method [34].All resulting constructs were verified by sequencing and all transgenic lines were confirmed by targeted genome sequencing and qRT-PCR.Primers of plasmid vector construction for genome editing and overexpression of SiPSY1 are listed in Table S2.
2.3.Structural and spatiotemporal expression pattern of SiPSY1
A phylogenic tree of homologous PSY proteins across cereal crops was constructed by MEGA5.0 [35].The promoter (3021 bp upstream)sequence of SiPSY1 was obtained by PCR and ligated into pCAMBIA1305.1 using an In-Fusion Advantage PCR Cloning Kit(Clontech), and the plasmid was transformed into calli variety Ci846 using Agrobacterium strain EHA105.Transgenic lines were confirmed by PCR and GUS staining.Germinated seeds, seedlings,leaves and panicles of T1transgenic individuals were identified by GUS staining protocols.Vector p16318hGFP was used for subcellular localization.
Three accessions with high yellow pigment content (Ci173,Ci364, Ci383) and three with low content (Ci157, Ci162, Ci236)(Table S3)were selected for assaying transcription levels in developing grains by a qRT-PCR approach using an Applied Bio Systems 7300 analyzer.SiActin was used as the control and the relative transcription level of SiPSY1 was calculated by the 2–ΔΔCTmethod[36].
The transactivation assays in N.benthamiana leaves were performed according to Sun et al.[37] to evaluate activities of different promoters identified by haplotype analysis.About a 3-kb SiPSY1 promoter of each haplotype was fused with the luciferase reporter (LUC) gene by Gateway reactions (Invitrogen) into vector pGWB35 to generate reporter construct SiPSY1pro:LUC [38].LUC signals were observed and analyzed 48 h after infiltration using the Night SHADE LB 985 system (Berthold, Bad Wildbad,Germany).
2.4.Selection of SiPSY1 and grain YPC during domestication and recent improvement in foxtail millet
Nucleotide diversity data for both S.italica [28] and S.viridis[39]were retrieved from the internet(https://www.ncgr.ac.cn/MilletHap1 and https://www.ncbi.nlm.nih.gov/bioproject/?term=Development %20of%20sequence-based%20community%20tools%20for%20Setaria%20viridis-a%20model%20genetic%20system%20for%20C4%20grasses),π value of genomic sequences surrounding SiPSY1 on chromosome 4 were calculated for detection of selection pressure of SiPSY1 between domesticated and wild Setaria gene pools.Seed of 35 S.viridis accessions mentioned in a previous study[40]were sampled for seed YPC measurement.A total of 186 improved foxtail millet varieties released during the last six decades in China reported in previous studies [41] were also sampled for mature seed YPC measurement.The variation in YPC during domestication and improvement of foxtail millet were then examined.Seeds used for YPC measurement of both S.viridis and improved foxtail millet varieties were harvested from field cultivation of relevant accessions during 2015 growth period at Beijing, China (39.9°N,116.3°E, altitude 35 m).
2.5.Haplotype diversity analysis and KASP marker development
One hundred and one accessions(Table S3)with diverse carotenoid contents were sampled for haplotype variation analysis of both Cis-regulatory and ORF regions of SiPSY1 following Sangersequencing.All SNPs and InDels were screened by DnaSP 5 [42]and aligned by MEGA 5[34].Relationships between different haplotypes were plotted through Network 4.0 software[43].Diagnostic SNPs for different haplotypes were identified and KASP primers were developed following standard KASP guidelines.The allelespecific primers were designed carrying the standard FAM (5′-AC CTGCATACAGTCACACACTTTC-3′)and VIC(5′-ACCTGCATACA GTCA CACACTTTG-3′) tails and with the targeted SNP at the 3′end.A common primer (5′-AGCCTTTTGCTCGTGTGTCTGCTAA-3′) was designed so that the total amplicon length was about 50 bp.The primer mixture comprised of 46 μL ddH2O,30 μL common primer(100 μmol L-1), and 12 μL of each tailed primer (100 μmol L-1).Assays were made in 384-well formats and set up as ~3 μL reactions (10–20 ng μL-1dry DNA, 3 μL of 1× KASP master mixture,and 0.056 μL of primer mixture).PCR cycling was performed using the following protocol: hot start at 94 °C for 15 min, followed by ten touchdown cycles(94°C for 20 s; touchdown at 61°C initially and decreasing by 0.6°C per cycle for 25 s,for 10 cycles),followed by 26 additional cycles of annealing(94°C for 10 s;55°C for 60 s).
3.Results
3.1.Genome-wide association analysis identified SiPSY1 as the candidate regulatory gene for yellow grain pigment in foxtail millet
The 390 accessions were sampled for yellow pigment, lutein and zeaxanthin measurement in two environments (Figs.1A–D,S2A and B).Normal distributions of all three compounds were identified(Fig.1B–D).BLUE values of grain yellow pigment content ranged from 1.78 to 33.34 mg kg-1, with an average of 19.75 mg kg-1; grain lutein content ranged from 0.29 to 11.58 mg kg-1with an average of 6.17 mg kg-1; grain zeaxanthin content ranged from 0.11 to 3.02 mg kg-1with 1.70 mg kg-1as an average value.GWAS analysis for YPC, lutein and zeaxanthin contents conducted using the BLUE (Fig.1E) values revealed a single significant locus (Chr.4: 40,036,791) with the highest statistical value across all nine chromosomes in foxtail millet(Fig.2A).Moreover,similar results were obtained in GWAS for each environment(Fig.S2C).Local genomic LD analysis using BLUE values surrounding the most significant SNP revealed a LD block (Chr.4:40,035,115–40,047,842) covering three annotated genes (Chr.4:40,036,791).According to the genomic annotations of the foxtail millet reference genome of Yugu 1, one transcribed gene (Seita.4G288600) encoding phytoene synthase (PSY) was identified as the SiPSY1candidate (Fig.2B).
3.2.Functional verification of SiPSY1 as the key regulator essential for yellow grain pigment in foxtail millet
To determine whether SiPSY1 played a vital role in yellow pigment accumulation in foxtail millet grain, the CRISPR/Cas9 vector designed for targeting the coding region of SiPSY1 and overexpression vector of SiPSY1 driven by the 35S promoter were transformed into cultivar Ci846 for identification of yellow pigment content in mature seeds (Fig.2 C–I).
Genome editing of SiPSY1 caused a deletion of a ‘T’ in the 3rd exon of SiPSY1, leading to a frame shift mutation at the 191st amino acid and resulting in a truncated protein (Fig.2C).There was a significant decrease in grain carotenoid content in genome edited lines (Fig.2D–F) with YPC, lutein and zeaxanthin contents respectively reduced by 58.3%, 73.4% and 63.2% relative to Ci846,and suggesting the essential function of SiPSY1 in yellow pigmentation in grains of foxtail millet.Overexpression of SiPSY1 caused a significant increase in YPC in mature seeds of the transformed line (Fig.2G–I).The grain YPC, lutein and zeaxanthin content of overexpressing line OE13 were respectively increased by 1.15-,1.50- and 1.16-fold compared with Ci846.
The SiPSY1 genomic region consisted of six exons and five introns, and encoded a 415 aa protein (Fig.3A).In foxtail millet,three homologous PSY genes (Seita.4G288600, Seita.3G397800, Seita.2G303000) were identified in the genomic annotation of the Yugu 1 reference genome (https://phytozome.jgi.doe.gov/pz/por tal.html#!info?alias=Org_Sitalica).Phylogenetic analysis of PSYs sourced from different crop species (Fig.3B) revealed that foxtail millet PSYs were more similar to PSYs derived from maize and sorghum.Sub-cellular studies of fused SiPSY1 and HGFP in foxtail millet protoplasts located SiPSY1 in the chloroplasts (Fig.3C),suggesting possible roles of SiPSY1 in chloroplast development.
3.3.Transcriptional variations in SiPSY1 at grains from 15 DAP determine yellow pigmentation accumulation in foxtail millet
To characterize the transcriptional pattern of SiPSY1 in foxtail millet, pSiPsy1::GUS was transformed into foxtail millet to detect expression of SiPSY1 in different tissues of developing foxtail millet plants(Fig.3D–L).GUS staining revealed that SiPSY1 was expressed in leaves, panicles and spikelets, but not in roots.To characterize the effect of SiPSY1 on yellow pigment accumulation in developing grains, pollinated spikelets were sampled from yellow and white grain accessions (Fig.3M).No obvious differences in transcriptional levels were identified during 0–10 DAP (days after pollination), but the expression level of SiPSY1 was significantly higher in the yellow accessions from 15 DAP (Fig.3M), suggesting transcriptional level of SiPSY1 was the key factor in determining yellow pigment level in foxtail millet.The sharp decrease in expression level of SiPSY1 after 15 DAP in the white grain foxtail millet accessions might contribute to their lower yellow pigmentation,and the genomic variation in Cis-regulating regions of SiPSY1 could be the main factor influencing grain color in this crop species.
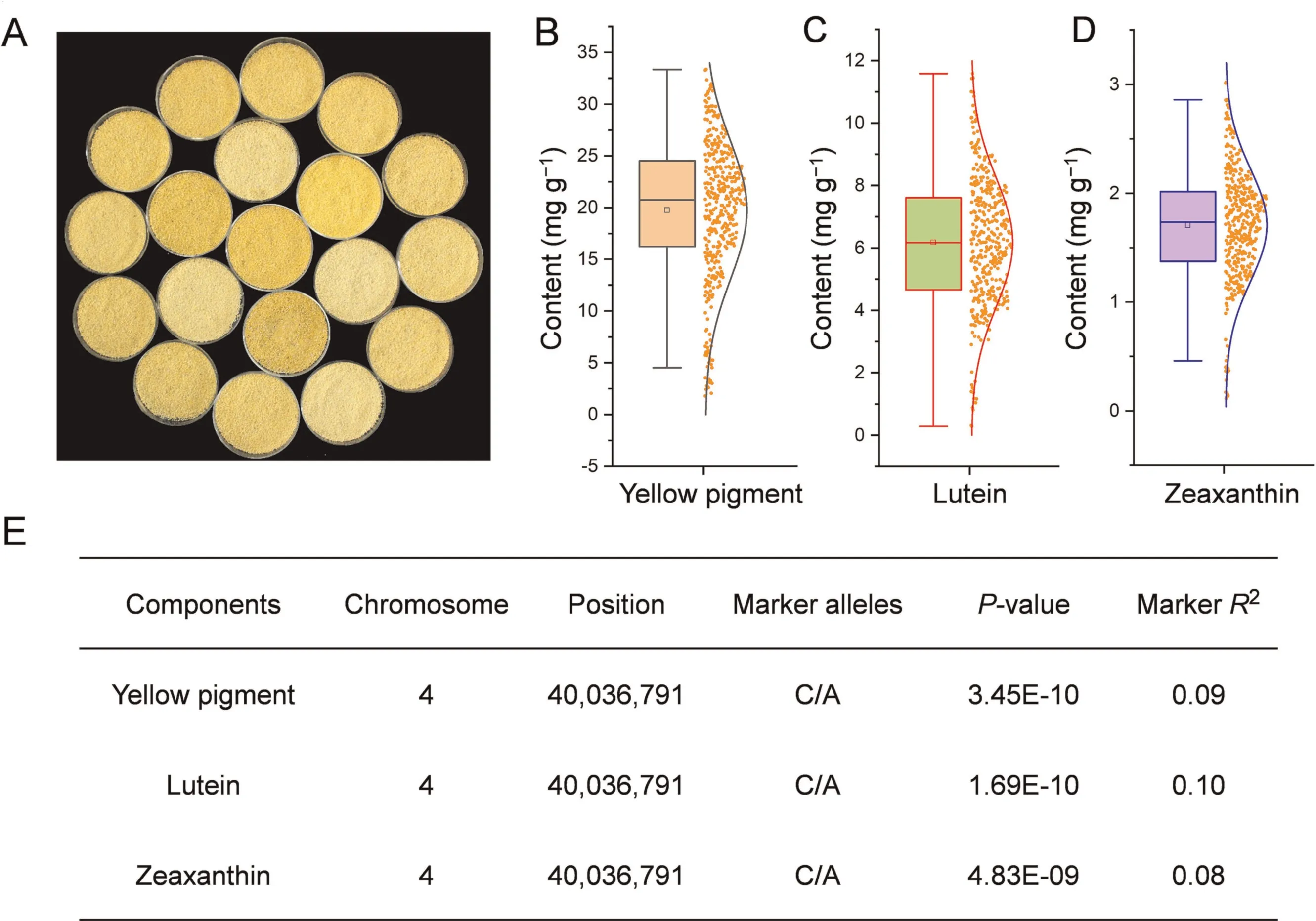
Fig.1.Characterization of yellow pigment, lutein and zeaxanthin contents in mature grains of foxtail millet accessions.(A) Milled grain samples of foxtail millet varieties showing different color intensities.(B–D) Distributions of BLUE values of three traits measured by flour samples derived from mature grains from two environments.(E)Significant loci identified for BLUE values of the three components by GWAS analysis.
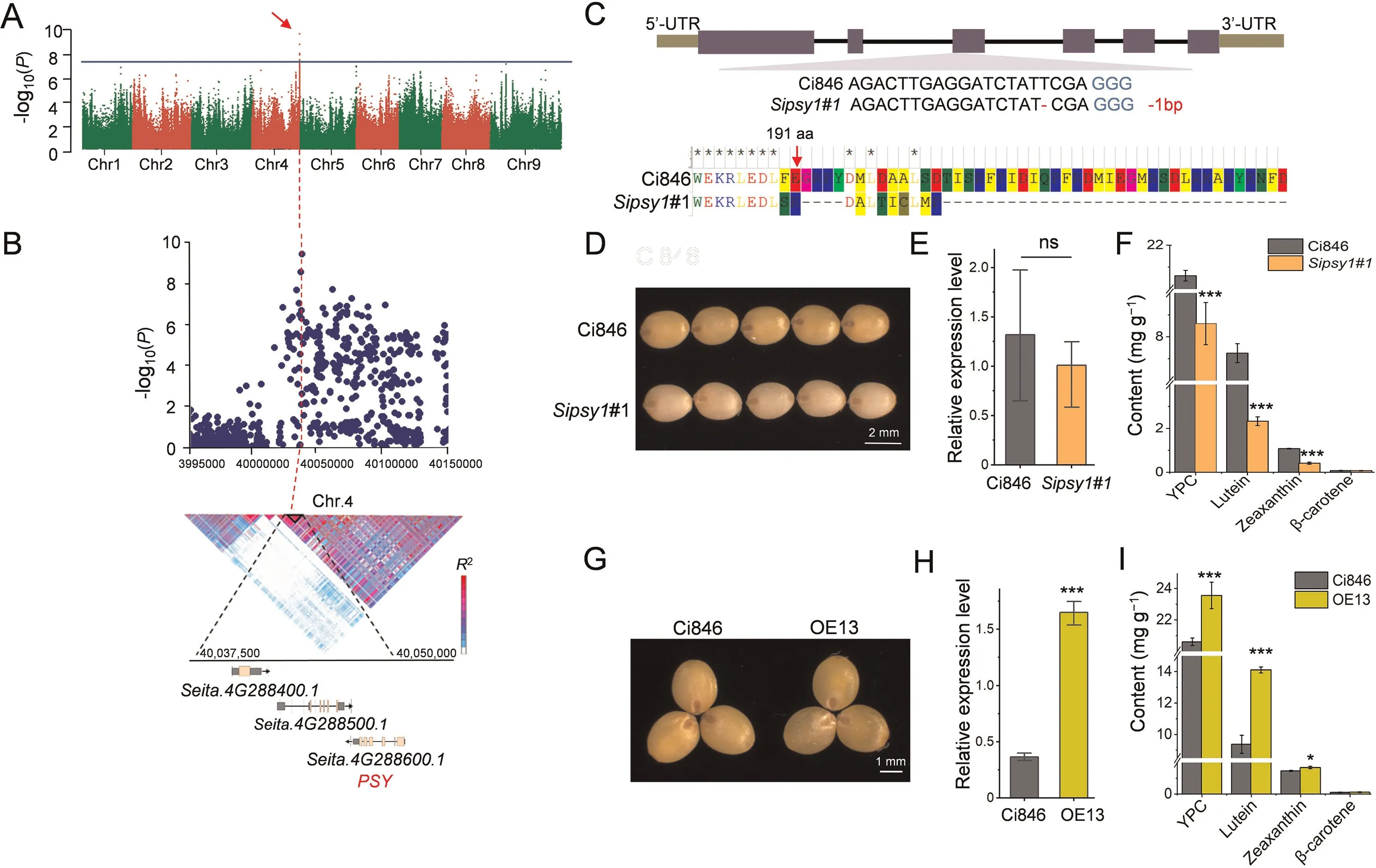
Fig.2.Genome-wide association analysis of yellow pigment content in foxtail millet grains and functional verification of SiPSY1 in foxtail millet by CRISPR/Cas9.(A)Wholegenome manhattan plot of yellow pigment accumulation in foxtail millet flour.Red arrow indicates the highest signal dot identified on chromosome 4.(B)Local manhattan plot and local LD block analysis of SiPSY1 and surrounding region.Black triangle represents the LD block closely associated with YPC.(C)Location of genome-edited site in the 3rd exon of SiPSY1 and mutation in amino acid 191 of SiPSY1.(D) Morphological characterizations of mature seeds of wild type Ci846 and genome edited lines; Scale bar,2 mm.(E)Transcription level of SiPSY1 CRISPR mutant relative to Ci846.(F)Contents of YPC,lutein,zeaxanthin and β-carotene in mature grains of the wild type(black)and genome modified (orange) individuals.(G) Morphological characterizations of mature seeds of wild type Ci846 and overexpression line, Scale bar, 1 mm.(H) Transcription levels of SiPSY1 overexpression lines.(I) Contents of YPC, lutein, zeaxanthin and β-carotene, in mature grains of both wild type and overexpression individuals.*, P < 0.05;***, P < 0.001; ns, no significance.

Fig.3.Genomic structure and expressional pattern of SiPSY1.(A)Genomic structure of SiPSY1.(B)Phylogenic relationships among PSY homologs sourced from different species.NCBI ID for all sequences were as follows: AAK07735.1 (Oryza sativa PSY1),XP_015618170.1 (Oryza sativa PSY2), XP_015611707.1(Oryza sativa PSY3),NP_001031895.1(Arabidopsis thaliana PSY1), AAB60314.1 (Zea mays PSY1), NP_001108117.1 (Zea mays PSY2), ABC75827.1 (Zea mays PSY3), XP_021305193.1 (Sorghum bicolor PSY1), XP_002462810.1 (Sorghum bicolor PSY3),ADZ24219.1(Nicotiana tabacum PSY1),NP_001312950.1(Nicotiana tabacum PSY2),NP_001234812.1(Solanum lycopersicum PSY1),P37273.1(Solanum lycopersicum PSY2),NP_001316106.1(Daucus carota PSY1),NP_001316096.1(Daucus carota PSY2),XP_017217851.1(Daucus carota PSY3),ACF42352.1(Triticum aestivum PSY1),AGL08127.1(Brassica napus PSY1),AGL08128.1(Brassica napus PSY2).(C)Subcellular localization of SiPSY1.Scale bar,10 μm.(D-L)GUS staining of germinated seed,leaves,panicles,floret and immature seeds.(D) Radicle, Scale bar,5 mm.(E)Seedling leaves.Scale bar,5 mm.(F) Leaf sheath and pulvinus.Scale bar,5 mm.(G)Stem.Scale bar,5 mm.(H) Spike-stalk.Scale bar,5 mm.(I)Primary branch of panicle.Scale bar,5 mm.(J)Floret.Scale bar,2 mm.Immature seeds at(K)10 DAP and(L)15 DAP.Scale bar,1 mm.(M)Transcriptional level of SiPSY1 during mature seed development after pollination in both yellow(high YPC)and white (low YPC)type foxtail millet accessions.**, P < 0.01 (Student’s t-test).
3.4.SiPSY1 was selected during domestication of Setaria spp.and is still selectable for future breeding of foxtail millet
To clarify whether SiPSY1 was involved in domestication or recent breeding of foxtail millet, genome sequence diversity of SiPSY1 in both wild S.viridis and cultivated S.italica accessions was determined, which suggested that SiPSY1 was localized in a genomic region that was under selection during domestication of foxtail millet(Fig.4A).The grain YPC of 35 accessions of wild Setaria viridis ranged from 1.16 mg kg-1to 8.30 mg kg-1with an average of 3.82 mg kg-1, whereas the grain YPC in 186 domesticated foxtail millet accessions ranged from 0.18 mg kg-1to 42.8 mg kg-1with an average of 19.5 mg kg-1(Fig.4B),indicating a significant change in grain YPC between the wild and domesticated species.
Yellow grain pigmentation has been a key quality trait in foxtail millet breeding programs in recent decades because of commercial preference.However, YPC characterization of cultivars released over the past six decades showed no significant increase in mean YPC.For instance, the average YPC of varieties released during the 1990s was 25.2 mg kg-1, whereas the average YPC of cultivars released during the 2000s was 24.2 mg kg-1(Fig.4C), indicating great potential for increasing YPC in future breeding programs.
3.5.Haplotype diversity is relevant to transcriptional variation in SiPSY1 among foxtail millet varieties
To identify natural genomic variation in the Cis-regulatory regions and effect of human selection on SiPSY1 in foxtail millet germplasm, a panel of 101 diverse foxtail millet accessions were sequenced.After screening a 2800 bp genomic region upstream of the SiPSY1 initiation codon,76 SNPs,2 InDels and 13 haplotypes were identified(Fig.5A).According to accession numbers that represent counterpart haplotypes in all sampled germplasm,the three most frequent haplotypes (hap_2, hap_4 and hap_5; Fig.5B) were selected for further analysis.
To clarify the relationship between YPC and activities of the three main haplotypes in the promoter region of SiPSY1, yellow pigment accumulation in foxtail millet grains were identified for all three haplotypes (Fig.6A).Hap_4 accessions possessed the highest level of yellow pigment(mean value 23.35±6.81 mg kg-1),followed by hap_2 (17.00 ± 6.25 mg kg-1) and hap_5 (3.76 ± 2.11 mg kg-1), indicating significant difference of yellow pigment among the three haplotypes.
Fluorescence analysis of tobacco leaves infused by both hap_5 and hap_4 activated the luciferase reporter (Fig.6B) and revealed higher intensity of fluorescence for hap_4(mean 3900 counts/second) compared with hap_5 (2100 counts/second).This suggested that the hap_4 promoter was stronger than hap_5 influencing yellow pigment accumulation in grain sourced from diverse foxtail millet accessions.These results indicated that selection of hap_4 in foxtail millet breeding programs should lead higher level of grain carotenoids.
3.6.Development of a KASP marker for higher levels of yellow pigment in foxtail millet grain
To develop diagnostic marker for high-throughput, costeffective screening of superior SiPSY1 promoters in foxtail millet breeding programs, a KASP assay was developed based on SNP(C/G) at position 2645 bp in promoter region of SiPSY1.The KASP assay identified genotypes with the cytosine to guanine transversion associated with higher levels of yellow pigment accumulation in the grain (Fig.6C).Sampling of 101 accessions to confirm the effect of the KASP assay on detecting genotypes with higher yellow grain pigment (Fig.6D).‘G’ genotypes had a yellow pigment accumulation of 17.5 mg kg-1compared to ‘C’ genotypes with a mean of 4.5 mg kg-1.These results suggest that the KASP assay(explaining 74.3% of total YPC variation) could be effective in identifying lines with higher yellow grain pigment in breeding programs.

Fig.4.Selection of SiPSY1 and yellow seed color during domestication and breeding of foxtail millet.(A)Selection of SiPSY1 inferred by genomic diversity analysis in both wild S.viridis accessions and cultivated S.italica landraces.(B) Variations in YPC in S.viridis (n = 35) sampled from different eco-regions and S.italica released during last six decades (n = 186) in China.(C) Variation in YPC in improved varieties released during the last six decades in China.***, P < 0.001; NS, not significant.
4.Discussion
4.1.PSY1 plays a vital role in carotenoid accumulation and could be an ideal gene for the enhancement of nutritional quality in cereal grains
Vitamin A deficiency(VAD)causes blindness in 250,000–500,000 children every year across the globe,with nearly half of them dying from VAD-related illnesses within several months[7].PSY1 has been proposed as a key regulator of the carotenoid(precursors of vitamin A)biosynthetic pathway in many crop species,including maize[14],wheat [44] and rice [2].This pathway has been modified to create genetically modified organisms such as golden rice for enhancement of nutritional value of rice in Africa[6].Although it was found that PSY2 could compensate for the absence of PSY1 in pepper fruit [9]such compensation between PSYs has not been confirmed in cereal crop species [2].In maize, there are three PSYs (PSY1, PSY2 and PSY3)but only PSY1 was confirmed to be associated with grain carotenoid accumulation [18].PSY2 is expressed in leaves [45] and PSY3 was induced in roots and confirmed to be involved in abiotic stress tolerance[46].Three PSYs(Fig.3B)were identified in foxtail millet in the present study and further studies are required to dissect functional diversity of SiPSYs in this model species to enable a better understanding of regulatory patterns of different PSYs in a range of plant species.In the recent published work of metabolite trait annotations in foxtail millet[25]visible color differences in foxtail millet grains evaluated by‘b*’was attributed to PSY1.In the present study,a genome-wide association study conducted in foxtail millet also revealed that PSY1 was the key regulatory factor in determining yellow pigment accumulation.Knockout and overexpression of SiPSY1 did dramatically change yellow pigment content in foxtail millet mature grains,consistent with the previously reported achievement in creation of golden rice[47].Map-based cloning of a white panicle mutant in foxtail millet also revealed that SiPSY1 had a role in panicle development [48].Phylogenetic analysis also suggested that the coding sequence and enzyme function of PSY1 is conserved across maize,rice and foxtail millet.Based on the present results,it seems that PSY1 is functionally conserved in cereal crop species,and could be selection target in other cereal crops,including rice,wheat,maize,sorghum,foxtail millet,common millet(Panicum miliaceum L.),pearl millet(Pennisetum americarum L.),finger millet(Eleusine coracana L.)and other millets.Cereals provide nearly 70% of total energy and nutrition for human beings and livestock; however, only maize,wheat,foxtail millet and common millet have evolved yellow grain with higher carotenoid contents.Therefore, deciphering of regulatory patterns of PSY1 from high carotenoid crops may assist in breeding cereals varieties to meet consumer demands.
4.2.Genomic variation in SiPSY1 revealed superior alleles for breeding of foxtail millet

Fig.5.Haplotype analysis of promoter region of SiPSY1.(A)Base sequences of 13 haplotypes identified in foxtail millet(‘*1’represents:TGGTGCAAGGCTTAGGCCTCCTGCGCCAATGCAAGTTAAGTGGTTTGAT; ‘*2’ represents: GTGCAGCTAGGCTCCTGCGCCAATGCAAGTAAGTGTTTGA; ‘IN1’ represents: ACGTGCCAAAAGGAAATTCAGCGTATATC).(B)Relationship and network of all haplotypes.Number of accessions with each haplotype is indicated by circle area.
Although it has been inferred that PSY1 is functionally conserved in cereal crop species,few studies have examined haplotype variation in PSY1 in cereal crops.The results of Fu et al.[17]based on maize, teosinte, tripsacum, coix, and sorghum indicated that PSY1 has the same gene structure across species.A similar genomic structure was also identified in foxtail millet(Fig.3A)in this study,further confirming the conserved PSY1 gene sequences during the evolution of grasses.Moreover, the large variation in carotenoid content identified among foxtail millet and maize accessions [17]also indicate genetic differentiation among carotenoid synthesisrelated genes.In this study, 78 polymorphic sites were identified in the cis-regulatory region of SiPSY1, indicating that SiPSY1 possessed a higher level of genomic variation with no positive selection identified in the gene coding region.This observation was consistent with previous results for many other cereal species[49], implying potential for selection of superior alleles in future cereal breeding programs.Moreover, PSY1 was selected during domestication of foxtail millet (Fig.4A) evidenced by significant differences in YPC between wild S.viridis and cultivated S.italica.However,the underlying mechanisms of increased YPC in the grain of foxtail millet needs further clarification.Seven SiPSY1 haplotypes were detected by high-throughput sequencing in previous reports [25], and in this study, 13 haplotypes were identified in the promoter region (Fig.5A, B).This implied abundant variation in SiPSY1 that allow selection in breeding programs.
To facilitate marker assisted selection,KASP assay based on a C/G SNP (Fig.6D) was developed for identification of a SiPSY1 favorable allele associated with high yellow pigment accumulation.However, other polymorphic sites in the cis-regulatory region could also be valuable for selection of specific rare alleles.
Sequence variations in the coding region of SiPSY1 were also identified (Fig.S2) and a single missense mutation (2327 bp) in exon 5 was associated with an amino acid change from phenylalanine to leucine(Table S1).Further study is required to clarify if this variation affects functionality of the protein.

Fig.6.Identification of superior promoter haplotypes and development of a KASP assay for superior allele identification of SiPSY1.(A) Mean yellow pigment contents in accessions belonging to three major haplotypes.(B)Transactivation assays of two haplotypes infused into N.benthamiana leaves.**,P<0.01(Student’s t-test).(C)Scatter plots of a KASP assay showing ‘‘G” on FAM (blue) and ‘‘C” (red) on VIC.(D) Effect of the KASP marker on detection of superior alleles.
4.3.Genomic modification of the carotenoid biosynthesis pathway is an effective approach for enhancing yellow pigment accumulation in foxtail millet
The carotenoid biosynthesis pathway was considered an important determinant of accumulation of kernel color in a previous genome-wide association mapping study [18] of regulators affecting kernel color in maize.In this study of foxtail millet,SiPSY1 was expressed at an early stage of grain development.This expression pattern was also identified in developing grains of wheat [44],implying conserved regulatory model of PSY1 in determination of carotenoid biosynthesis in developing grains of cereal plants.Sub-cellular localization and spatio-temporal transcription analysis also suggested that SiPSY1 was functionally conserved[2]across cereal crop species.In terms of other regulators relevant to carotenoid accumulation in cereal grains, a rare natural variation of crtRB1 gene in maize carotenoid biosynthesis pathway has been confirmed also influence β-carotene content of maize grain [7],which implied that foxtail millet homologous crtRB1 might possess similar function and could be genetically modified for enhancement of yellow pigment accumulation in Setaria.
In this study,the expression pattern of SiPSY1 in immature grains revealed significant difference between yellow and white kernel accessions from 15 DAP.This might determine carotenoids biosynthesis in foxtail millet varieties with different grain yellow pigment content,which would mainly accumulate after 15 DAP in pollinated kernels of foxtail millet.An interesting result of dramatically declined transcription level(Fig.3M)of SiPSY1 in white grain accessions was observed, which may be due to negative regulators appeared at 15 DAP interrupt expression of SiPSY1.The SIFGD [50]database was used to search for microRNAs targeting SiPSY1 in foxtail millet,but no reported microRNA closely linked with or targeting SiPSY1 in foxtail millet was identified, implying more efforts are needed in future to clarify negative regulators associated with this interesting observation.In a previous study on yellow pigment accumulation trends during grain ripening in foxtail millet [51], a rapid decrease in yellow pigment content was due to delayed harvesting after grain ripening.Given that transcription level of SiPSY1 was decreased 15 DAP during early development of pollinated grains in foxtail millet (Fig.3M), with abrupt fall in the white grain genotype but a gradual decrease in the yellow genotype,it can be inferred that homeostasis of biosynthetic and metabolic of carotenoid in foxtail millet might be the causation of decreased yellow pigment accompany with delayed harvesting after ripening.Similar decreased transcription level of PSY1 was also reported in 21 DAP kernels of wheat[44],implied that homeostasis of PSY1 associated with kernel ripening in cereal crops might be conserved.However, this hypothesis requires further investigation in future.
5.Conclusions
In this study, a genomic region associated with yellow grain pigment in grains of foxtail millet was identified by genomewide association analysis, and the SiPSY1 gene was confirmed as the main contributor to yellow pigment accumulation in this species.Transcription of SiPSY1 over the first 15 days after pollination was responsible for the accumulation of yellow pigment in the seeds.Expression of SiPSY1 in white seeded genotypes fell abruptly from 15 DAP whereas in yellow seeded genotypes there was a gradual decline but at a higher level of pigment concentration.Haplotype analyses covering both promoter and CDS regions of SiPSY1 were conducted, and a KASP assay based on a SNP in the upstream cis-regulatory region was developed for marker assisted selection of a favorable yellow allele.Results of this work will promote genetic dissection of nutritional characters in foxtail millet to enhance quality traits of this ancient crop species.
CRediT authorship contribution statement
Rongjun Zuo:Investigation,Formal analysis,Writing–original draft.Yanyan Zhang:Investigation, Formal analysis.Yanbing Yang:Investigation.Chunfang Wang:Investigation.Hui Zhi:Investigation.Linlin Zhang:Investigation.Sha Tang:Investigation.Yanan Guan:Formal analysis.Shunguo Li:Investigation.Ruhong Cheng:Formal analysis.Zhonglin Shang:Formal analysis.Guanqing Jia:Supervision, Investigation, Formal analysis, Writing –original draft.Xianmin Diao:Supervision,Writing–original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China(U21A20216),the National Key Research and Development Program of China (2018YFD1000705/2018YFD1000700),Fundamental Research Funds of the Chinese Academy of Agricultural Sciences(Grant to Guanqing Jia,1610092016116,Y2017JC15),China Agricultural Research System (CARS06-14.5-A04), State Key Laboratory of Crop Gene Resources and Breeding, Key Laboratory of Grain Crop Genetic Resources Evaluation and Utilization, Key Laboratory of Crop Gene Resource and Germplasm Enhancement (MOA), and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.09.008.
- The Crop Journal的其它文章
- OsSPL10 controls trichome development by interacting with OsWOX3B at both transcription and protein levels in rice (Oryza sativa L.)
- Ectopic expression of OsNF-YA8, an endosperm-specific nuclear factor Y transcription-factor gene, causes vegetative and reproductive development defects in rice
- Mechanisms of autophagy function and regulation in plant growth,development, and response to abiotic stress
- ZmDRR206 functions in maintaining cell wall integrity during maize seedling growth and defense response to external stresses
- The plasmodesmata-associated β-1,3-glucanase gene GhPdBG regulates fiber development in cotton
- The MabHLH11 transcription factor interacting with MaMYB4 acts additively in increasing plant scopolin biosynthesis

